Role of lysine binding residues in the global folding of the lysC riboswitch
- PMID: 26403229
- PMCID: PMC4829333
- DOI: 10.1080/15476286.2015.1094603
Role of lysine binding residues in the global folding of the lysC riboswitch
Abstract
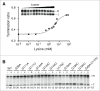
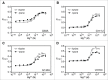
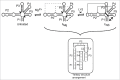
Riboswitches regulate gene expression by rearranging their structure upon metabolite binding. The lysine-sensing lysC riboswitch is a rare example of an RNA aptamer organized around a 5-way helical junction in which ligand binding is performed exclusively through nucleotides located at the junction core. We have probed whether the nucleotides involved in ligand binding play any role in the global folding of the riboswitch. As predicted, our findings indicate that ligand-binding residues are critical for the lysine-dependent gene regulation mechanism. We also find that these residues are not important for the establishment of key magnesium-dependent tertiary interactions, suggesting that folding and ligand recognition are uncoupled in this riboswitch for the formation of specific interactions. However, FRET assays show that lysine binding results in an additional conformational change, indicating that lysine binding may also participate in a specific folding transition. Thus, in contrast to helical junctions being primary determinants in ribozymes and rRNA folding, we speculate that the helical junction of the lysine-sensing lysC riboswitch is not employed as structural a scaffold to direct global folding, but rather has a different role in establishing RNA-ligand interactions required for riboswitch regulation. Our work suggests that helical junctions may adopt different functions such as the coordination of global architecture or the formation of specific ligand binding site.
Keywords: RNA folding; Riboswitch; helical junction; lysine; metabolite-sensing.
Figures



References
-
- Serganov A, Nudler E. A decade of riboswitches. Cell 2013; 152:17-24; PMID:23332744; http://dx.doi.org/ 10.1016/j.cell.2012.12.024 - DOI - PMC - PubMed
-
- Edwards TE, Klein DJ, Ferre-D'Amare AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol 2007; 17:273-9; PMID:17574837; http://dx.doi.org/ 10.1016/j.sbi.2007.05.004 - DOI - PubMed
-
- Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angewandte Chemie 2007; 46:1212-9; PMID:17226886; http://dx.doi.org/ 10.1002/anie.200604163 - DOI - PubMed
-
- Haller A, Souliere MF, Micura R. The Dynamic Nature of RNA as Key to Understanding Riboswitch Mechanisms. Acc Chem Res 2011:1339-48; PMID:21678902; http://dx.doi.org/ 10.1021/ar200035g - DOI - PubMed
-
- Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 2007; 8:R239; PMID:17997835; http://dx.doi.org/ 10.1186/gb-2007-8-11-r239 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
