High-throughput matrix screening identifies synergistic and antagonistic antimalarial drug combinations
- PMID: 26403635
- PMCID: PMC4585899
- DOI: 10.1038/srep13891
High-throughput matrix screening identifies synergistic and antagonistic antimalarial drug combinations
Abstract
Drug resistance in Plasmodium parasites is a constant threat. Novel therapeutics, especially new drug combinations, must be identified at a faster rate. In response to the urgent need for new antimalarial drug combinations we screened a large collection of approved and investigational drugs, tested 13,910 drug pairs, and identified many promising antimalarial drug combinations. The activity of known antimalarial drug regimens was confirmed and a myriad of new classes of positively interacting drug pairings were discovered. Network and clustering analyses reinforced established mechanistic relationships for known drug combinations and identified several novel mechanistic hypotheses. From eleven screens comprising >4,600 combinations per parasite strain (including duplicates) we further investigated interactions between approved antimalarials, calcium homeostasis modulators, and inhibitors of phosphatidylinositide 3-kinases (PI3K) and the mammalian target of rapamycin (mTOR). These studies highlight important targets and pathways and provide promising leads for clinically actionable antimalarial therapy.
Figures

 = Endoperoxides;
= Endoperoxides;  = HLF, LUM, MFQ, CQ, TFQ;
= HLF, LUM, MFQ, CQ, TFQ;  = hPI3K/mTOR;
= hPI3K/mTOR;  = mitochondrial/DHODH;
= mitochondrial/DHODH;  = ion channel modulator;
= ion channel modulator;  = hybrid mechanism;
= hybrid mechanism;  = other). Vector lengths do not reflect the strength or weakness of the interaction. Additional network plots reflecting antagonistic outcomes are found in the SI. Enlarged versions of this figure can be found online at
= other). Vector lengths do not reflect the strength or weakness of the interaction. Additional network plots reflecting antagonistic outcomes are found in the SI. Enlarged versions of this figure can be found online at  = antimalarial drugs;
= antimalarial drugs;  = growth inhibitor;
= growth inhibitor;  = hPI3K/mTOR;
= hPI3K/mTOR;  = mitochondrial/DHODH;
= mitochondrial/DHODH;  = ion channel modulator;
= ion channel modulator;  = signaling/transporter inhibitor. Large symbols correspond to the high potency class and small symbols correspond to medium potency class). (D) Hierarchical clustering of combination profiles based upon 1) synergy assessment; 2) potency class; 3) mechanism of action (MOA) relationship. The number of clusters (six) was selected based on the largest number of clusters that led to zero or one cluster that was not enriched in any MOA combination (at the 0.01 level). (E) A summary of MOA combinations enriched in cluster 3 (green), relative to the entire dataset (see SI for full list of code definitions). Enrichment was tested using Fisher’s exact test with p-values adjusted using the Benjamini-Hochberg method, and plotted as –log10 p-value. The blue and red dashed lines correspond to p = 0.05 and p = 0.01 respectively. While six MOA combinations were significant at the 0.01 level, we annotated the top three. See SI for full figure.
= signaling/transporter inhibitor. Large symbols correspond to the high potency class and small symbols correspond to medium potency class). (D) Hierarchical clustering of combination profiles based upon 1) synergy assessment; 2) potency class; 3) mechanism of action (MOA) relationship. The number of clusters (six) was selected based on the largest number of clusters that led to zero or one cluster that was not enriched in any MOA combination (at the 0.01 level). (E) A summary of MOA combinations enriched in cluster 3 (green), relative to the entire dataset (see SI for full list of code definitions). Enrichment was tested using Fisher’s exact test with p-values adjusted using the Benjamini-Hochberg method, and plotted as –log10 p-value. The blue and red dashed lines correspond to p = 0.05 and p = 0.01 respectively. While six MOA combinations were significant at the 0.01 level, we annotated the top three. See SI for full figure.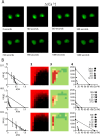

References
-
- WHO. Guidelines for the treatment of malaria. World Health Organization: Geneva, Switzerland (2010).
-
- Kuhn S., Gill M. J. & Kain K. C. Emergence of atovaquone-proguanil resistance during treatment of Plasmodium falciparum malaria acquired by a non-immune north American traveller to west Africa. Am. J. Trop. Med. Hyg. 72, 407–409 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

