A Novel Sucrose-Regulatory MADS-Box Transcription Factor GmNMHC5 Promotes Root Development and Nodulation in Soybean (Glycine max [L.] Merr.)
- PMID: 26404246
- PMCID: PMC4613224
- DOI: 10.3390/ijms160920657
A Novel Sucrose-Regulatory MADS-Box Transcription Factor GmNMHC5 Promotes Root Development and Nodulation in Soybean (Glycine max [L.] Merr.)
Abstract
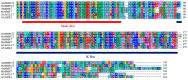
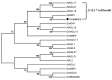
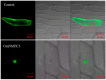
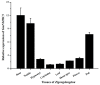
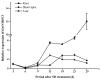
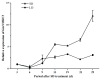
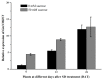
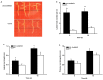
The MADS-box protein family includes many transcription factors that have a conserved DNA-binding MADS-box domain. The proteins in this family were originally recognized to play prominent roles in floral development. Recent findings, especially with regard to the regulatory roles of the AGL17 subfamily in root development, have greatly broadened their known functions. In this study, a gene from soybean (Glycine max [L.] Merr.), GmNMHC5, was cloned from the Zigongdongdou cultivar and identified as a member of the AGL17 subfamily. Real-time fluorescence quantitative PCR analysis showed that GmNMHC5 was expressed at much higher levels in roots and nodules than in other organs. The activation of expression was first examined in leaves and roots, followed by shoot apexes. GmNMHC5 expression levels rose sharply when the plants were treated under short-day conditions (SD) and started to pod, whereas low levels were maintained in non-podding plants under long-day conditions (LD). Furthermore, overexpression of GmNMHC5 in transgenic soybean significantly promoted lateral root development and nodule building. Moreover, GmNMHC5 is upregulated by exogenous sucrose. These results indicate that GmNMHC5 can sense the sucrose signal and plays significant roles in lateral root development and nodule building.
Keywords: Glycine max; GmNMHC5; MADS-box protein; lateral roots development; nodule building; sucrose.
Figures

Similar articles
-
In situ expression of the GmNMH7 gene is photoperiod-dependent in a unique soybean (Glycine max [L.] Merr.) flowering reversion system.Planta. 2006 Mar;223(4):725-35. doi: 10.1007/s00425-005-0130-y. Epub 2005 Oct 6. Planta. 2006. PMID: 16208488
-
GmEXPB2, a Cell Wall β-Expansin, Affects Soybean Nodulation through Modifying Root Architecture and Promoting Nodule Formation and Development.Plant Physiol. 2015 Dec;169(4):2640-53. doi: 10.1104/pp.15.01029. Epub 2015 Oct 2. Plant Physiol. 2015. PMID: 26432877 Free PMC article.
-
Promoters of orthologous Glycine max and Lotus japonicus nodulation autoregulation genes interchangeably drive phloem-specific expression in transgenic plants.Mol Plant Microbe Interact. 2007 Jul;20(7):769-80. doi: 10.1094/MPMI-20-7-0769. Mol Plant Microbe Interact. 2007. PMID: 17601165
-
MADS: the missing link between identity and growth?Trends Plant Sci. 2011 Feb;16(2):89-97. doi: 10.1016/j.tplants.2010.11.003. Epub 2010 Dec 7. Trends Plant Sci. 2011. PMID: 21144794 Review.
-
MADS-box genes underground becoming mainstream: plant root developmental mechanisms.New Phytol. 2019 Aug;223(3):1143-1158. doi: 10.1111/nph.15793. Epub 2019 Apr 12. New Phytol. 2019. PMID: 30883818 Review.
Cited by
-
GmNMHC5, A Neoteric Positive Transcription Factor of Flowering and Maturity in Soybean.Plants (Basel). 2020 Jun 25;9(6):792. doi: 10.3390/plants9060792. Plants (Basel). 2020. PMID: 32630379 Free PMC article.
-
GsMAS1 Encoding a MADS-box Transcription Factor Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana.Int J Mol Sci. 2020 Mar 15;21(6):2004. doi: 10.3390/ijms21062004. Int J Mol Sci. 2020. PMID: 32183485 Free PMC article.
-
The impact of the rhizobia-legume symbiosis on host root system architecture.J Exp Bot. 2020 Jun 26;71(13):3902-3921. doi: 10.1093/jxb/eraa198. J Exp Bot. 2020. PMID: 32337556 Free PMC article. Review.
-
Comparative transcriptome analysis of flower bud transition and functional characterization of EjAGL17 involved in regulating floral initiation in loquat.PLoS One. 2020 Oct 8;15(10):e0239382. doi: 10.1371/journal.pone.0239382. eCollection 2020. PLoS One. 2020. PMID: 33031442 Free PMC article.
-
Candidate genes expression profiling during wilting in chickpea caused by Fusarium oxysporum f. sp. ciceris race 5.PLoS One. 2019 Oct 23;14(10):e0224212. doi: 10.1371/journal.pone.0224212. eCollection 2019. PLoS One. 2019. PMID: 31644597 Free PMC article.
References
-
- Dong Z. Soybean Yield Physiology. Agricultural Press; Beijing, China: 2000. pp. 192–195.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

