T-cell receptor profiling in cancer
- PMID: 26404496
- PMCID: PMC5528728
- DOI: 10.1016/j.molonc.2015.09.003
T-cell receptor profiling in cancer
Abstract
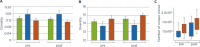
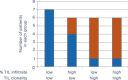
Immunosequencing is a platform technology that allows the enumeration, specification and quantification of each and every B- and/or T-cell in any biologic sample of interest. Thus, it provides an assessment of the level and distribution of all the clonal lymphocytes in any sample, and allows "tracking" of a single clone or multiple clones of interest over time or from tissue to tissue within a given patient. It is based on bias-controlled multiplex PCR and high-throughput sequencing, and it is highly accurate, standardized, and sensitive. In this review, we provide evidence that immunosequencing is becoming an important analytic tool for the emerging field of immune-oncology, and describe several applications of this approach, including the assessment of residual disease post therapy in lymphoid malignancies, the prediction of response to immunotherapeutics of solid tumors containing tumor infiltrating lymphocytes, the identification of clonal responses in vaccination, infectious disease, bone marrow reconstitution, and autoimmunity, and the exploration of whether there are population-based stereotyped responses to certain exposures or interventions.
Keywords: Ig/TCR sequence biomarkers; Immunosequencing; Lymphoid clonality assessment/tracking.
Copyright © 2015 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Bertness, V. , Kirsch, I. , Hollis, G. , Johnson, B. , Bunn, P.A. , 1985. T-cell receptor gene rearrangements as clinical markers of human T-cell lymphomas. N. Engl. J. Med. 313, 534–538. - PubMed
-
- Bunge, J. , Fitzpatrick, M. , 1993. Estimating the number of species: a review. J. Am. Stat. Assoc. 88, 364–373.
-
- Carlson, C.S. , Emerson, R.O. , Sherwood, A.M. , Desmarais, C. , Chung, M.W. , Parsons, J.M. , Steen, M.S. , LaMadrid-Herrmannsfeldt, M.A. , Williamson, D.W. , Livingston, R.J. , Wu, D. , Wood, B.L. , Rieder, M.J. , Robins, H. , 2013. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 4, 2680 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

