Assembling a Correctly Folded and Functional Heptahelical Membrane Protein by Protein Trans-splicing
- PMID: 26405032
- PMCID: PMC4646914
- DOI: 10.1074/jbc.M115.681205
Assembling a Correctly Folded and Functional Heptahelical Membrane Protein by Protein Trans-splicing
Abstract
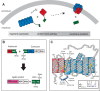
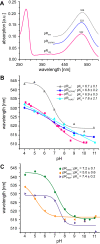
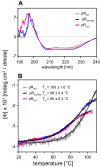
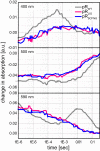
Protein trans-splicing using split inteins is well established as a useful tool for protein engineering. Here we show, for the first time, that this method can be applied to a membrane protein under native conditions. We provide compelling evidence that the heptahelical proteorhodopsin can be assembled from two separate fragments consisting of helical bundles A and B and C, D, E, F, and G via a splicing site located in the BC loop. The procedure presented here is on the basis of dual expression and ligation in vivo. Global fold, stability, and photodynamics were analyzed in detergent by CD, stationary, as well as time-resolved optical spectroscopy. The fold within lipid bilayers has been probed by high field and dynamic nuclear polarization-enhanced solid-state NMR utilizing a (13)C-labeled retinal cofactor and extensively (13)C-(15)N-labeled protein. Our data show unambiguously that the ligation product is identical to its non-ligated counterpart. Furthermore, our data highlight the effects of BC loop modifications onto the photocycle kinetics of proteorhodopsin. Our data demonstrate that a correctly folded and functionally intact protein can be produced in this artificial way. Our findings are of high relevance for a general understanding of the assembly of membrane proteins for elucidating intramolecular interactions, and they offer the possibility of developing novel labeling schemes for spectroscopic applications.
Keywords: intein; membrane protein; photoreceptor; protein folding; protein splicing; proton pump; seven-helix receptor; solid-state NMR.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Volkmann G., and Iwaï H. (2010) Protein trans-splicing and its use in structural biology: opportunities and limitations. Mol. Biosyst. 6, 2110–2121 - PubMed
-
- Yang J.-Y., and Yang W. Y. (2009) Site-Specific Two-Color Protein Labeling for FRET Studies Using Split Inteins. J. Am. Chem. Soc. 131, 11644–11645 - PubMed
-
- Ozawa T., and Umezawa Y. (2001) Detection of protein-protein interactions in vivo based on protein splicing. Curr. Opin. Chem. Biol. 5, 578–583 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

