Regulation of multispanning membrane protein topology via post-translational annealing
- PMID: 26408961
- PMCID: PMC4635508
- DOI: 10.7554/eLife.08697
Regulation of multispanning membrane protein topology via post-translational annealing
Abstract
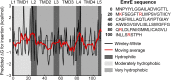
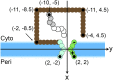
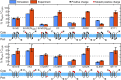
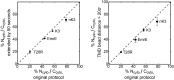
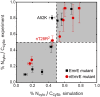
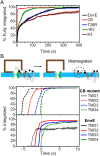
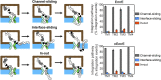
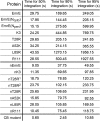
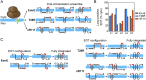
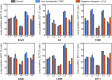
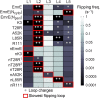
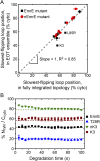
The canonical mechanism for multispanning membrane protein topogenesis suggests that protein topology is established during cotranslational membrane integration. However, this mechanism is inconsistent with the behavior of EmrE, a dual-topology protein for which the mutation of positively charged loop residues, even close to the C-terminus, leads to dramatic shifts in its topology. We use coarse-grained simulations to investigate the Sec-facilitated membrane integration of EmrE and its mutants on realistic biological timescales. This work reveals a mechanism for regulating membrane-protein topogenesis, in which initially misintegrated configurations of the proteins undergo post-translational annealing to reach fully integrated multispanning topologies. The energetic barriers associated with this post-translational annealing process enforce kinetic pathways that dictate the topology of the fully integrated proteins. The proposed mechanism agrees well with the experimentally observed features of EmrE topogenesis and provides a range of experimentally testable predictions regarding the effect of translocon mutations on membrane protein topogenesis.
Keywords: biochemistry; biophysics; dual-topology; membrane protein topology; none; simulation; structural biology; topogenesis.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



Comment in
-
The messy process of guiding proteins into membranes.Elife. 2015 Nov 6;4:e12100. doi: 10.7554/eLife.12100. Elife. 2015. PMID: 26544679 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

