Control of Chromatin Structure by Long Noncoding RNA
- PMID: 26410408
- PMCID: PMC4584417
- DOI: 10.1016/j.tcb.2015.07.002
Control of Chromatin Structure by Long Noncoding RNA
Abstract
Long noncoding RNA (lncRNA) is a pivotal factor regulating various aspects of genome activity. Genome regulation via DNA methylation and post-translational histone modifications is a well-documented function of lncRNA in plants, fungi, and animals. Here, we summarize evidence showing that lncRNA also controls chromatin structure, including nucleosome positioning and chromosome looping. We focus on data from plant experimental systems, discussed in the context of other eukaryotes. We explain the mechanisms of lncRNA-controlled chromatin remodeling and the implications of the functional interplay between noncoding transcription and several different chromatin remodelers. We propose that the unique properties of RNA make it suitable for controlling chromatin modifications and structure.
Keywords: RNA Polymerase V; chromatin remodeling; chromosome looping; nucleosome.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

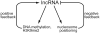
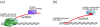
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

