Combined strategies for improving expression of Citrobacter amalonaticus phytase in Pichia pastoris
- PMID: 26410558
- PMCID: PMC4584009
- DOI: 10.1186/s12896-015-0204-2
Combined strategies for improving expression of Citrobacter amalonaticus phytase in Pichia pastoris
Abstract
Background: Phytase is used as an animal feed additive that degrades phytic acid and reduces feeding costs and pollution caused by fecal excretion of phosphorus. Some phytases have been expressed in Pichia pastoris, among which the phytase from Citrobacter amalonaticus CGMCC 1696 had high specific activity (3548 U/mg). Improvement of the phytase expression level will contribute to facilitate its industrial applications.
Methods: To improve the phytase expression, we use modification of P AOX1 and the α-factor signal peptide, increasing the gene copy number, and overexpressing HAC1 (i) to enhance folding and secretion of the protein in the endoplasmic reticulum. The genetic stability and fermentation in 10-L scaled-up fed-batch fermenter was performed to prepare for the industrial production.
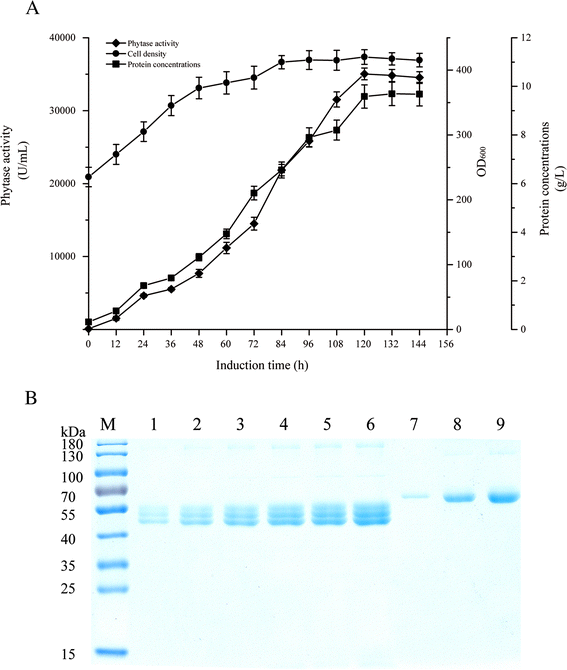
Results: The phytase gene from C. amalonaticus CGMCC 1696 was cloned under the control of the AOX1 promoter (P AOX1 ) and expressed in P. pastoris. The phytase activity achieved was 414 U/mL. Modifications of P AOX1 and the α-factor signal peptide increased the phytase yield by 35 and 12%, respectively. Next, on increasing the copy number of the Phy gene to six, the phytase yield was 141% higher than in the strain containing only a single gene copy. Furthermore, on overexpression of HAC1 (i) (i indicating induced), a gene encoding Hac1p that regulates the unfolded protein response, the phytase yield achieved was 0.75 g/L with an activity of 2119 U/mL, 412% higher than for the original strain. The plasmids in this high-phytase expression strain were stable during incubation at 30 °C in Yeast Extract Peptone Dextrose (YPD) Medium. In a 10-L scaled-up fed-batch fermenter, the phytase yield achieved was 9.58 g/L with an activity of 35,032 U/mL.
Discussion: The production of a secreted protein will reach its limit at a specific gene copy number where further increases in transcription and translation due to the higher abundance of gene copies will not enhance the secretion process any further. Enhancement of protein folding in the ER can alleviate bottlenecks in the folding and secretion pathways during the overexpression of heterologous proteins in P. pastoris.
Conclusions: Using modification of P AOX1 and the α-factor signal peptide, increasing the gene copy number, and overexpressing HAC1 (i) to enhance folding and secretion of the protein in the endoplasmic reticulum, we have successfully increased the phytase yield 412% relative to the original strain. In a 10-L fed-batch fermenter, the phytase yield achieved was 9.58 g/L with an activity of 35,032 U/mL. Large-scale production of phytase can be applied towards different biocatalytic and feed additive applications.
Figures




References
-
- Augspurger N, Webel D, Lei X, Baker D. Efficacy of an E. coli phytase expressed in yeast for releasing phytate-bound phosphorus in young chicks and pigs. J Anim Sci. 2003;81(2):474–483. - PubMed
-
- Chen C-C, Wu P-H, Huang C-T, Cheng K-J. A Pichia pastoris fermentation strategy for enhancing the heterologous expression of an Escherichia coli phytase. Enzym Microb Technol. 2004;35(4):315–320. doi: 10.1016/j.enzmictec.2004.05.007. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

