AlloRep: A Repository of Sequence, Structural and Mutagenesis Data for the LacI/GalR Transcription Regulators
- PMID: 26410588
- PMCID: PMC4783282
- DOI: 10.1016/j.jmb.2015.09.015
AlloRep: A Repository of Sequence, Structural and Mutagenesis Data for the LacI/GalR Transcription Regulators
Abstract
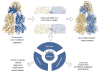
Protein families evolve functional variation by accumulating point mutations at functionally important amino acid positions. Homologs in the LacI/GalR family of transcription regulators have evolved to bind diverse DNA sequences and allosteric regulatory molecules. In addition to playing key roles in bacterial metabolism, these proteins have been widely used as a model family for benchmarking structural and functional prediction algorithms. We have collected manually curated sequence alignments for >3000 sequences, in vivo phenotypic and biochemical data for >5750 LacI/GalR mutational variants, and noncovalent residue contact networks for 65 LacI/GalR homolog structures. Using this rich data resource, we compared the noncovalent residue contact networks of the LacI/GalR subfamilies to design and experimentally validate an allosteric mutant of a synthetic LacI/GalR repressor for use in biotechnology. The AlloRep database (freely available at www.AlloRep.org) is a key resource for future evolutionary studies of LacI/GalR homologs and for benchmarking computational predictions of functional change.
Keywords: allostery; contact map; sequence alignment; transcription regulation; variant database.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
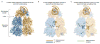

Similar articles
-
Data on publications, structural analyses, and queries used to build and utilize the AlloRep database.Data Brief. 2016 Jul 9;8:948-57. doi: 10.1016/j.dib.2016.07.006. eCollection 2016 Sep. Data Brief. 2016. PMID: 27508249 Free PMC article.
-
Comparing the functional roles of nonconserved sequence positions in homologous transcription repressors: implications for sequence/function analyses.J Mol Biol. 2010 Jan 29;395(4):785-802. doi: 10.1016/j.jmb.2009.10.001. Epub 2009 Oct 8. J Mol Biol. 2010. PMID: 19818797 Free PMC article.
-
Subdividing repressor function: DNA binding affinity, selectivity, and allostery can be altered by amino acid substitution of nonconserved residues in a LacI/GalR homologue.Biochemistry. 2008 Aug 5;47(31):8058-69. doi: 10.1021/bi800443k. Epub 2008 Jul 11. Biochemistry. 2008. PMID: 18616293 Free PMC article.
-
Allostery in the LacI/GalR family: variations on a theme.Curr Opin Microbiol. 2009 Apr;12(2):129-37. doi: 10.1016/j.mib.2009.01.009. Epub 2009 Mar 5. Curr Opin Microbiol. 2009. PMID: 19269243 Free PMC article. Review.
-
Flexibility and Disorder in Gene Regulation: LacI/GalR and Hox Proteins.J Biol Chem. 2015 Oct 9;290(41):24669-77. doi: 10.1074/jbc.R115.685032. Epub 2015 Sep 4. J Biol Chem. 2015. PMID: 26342073 Free PMC article. Review.
Cited by
-
Dynamics-based protein network features accurately discriminate neutral and rheostat positions.Biophys J. 2024 Oct 15;123(20):3612-3626. doi: 10.1016/j.bpj.2024.09.013. Epub 2024 Sep 13. Biophys J. 2024. PMID: 39277794
-
Nitrogen Fixation and Molecular Oxygen: Comparative Genomic Reconstruction of Transcription Regulation in Alphaproteobacteria.Front Microbiol. 2016 Aug 26;7:1343. doi: 10.3389/fmicb.2016.01343. eCollection 2016. Front Microbiol. 2016. PMID: 27617010 Free PMC article.
-
PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.Database (Oxford). 2023 May 3;2023:baad030. doi: 10.1093/database/baad030. Database (Oxford). 2023. PMID: 37171062 Free PMC article.
-
Deep representation learning improves prediction of LacI-mediated transcriptional repression.Proc Natl Acad Sci U S A. 2021 Jul 6;118(27):e2022838118. doi: 10.1073/pnas.2022838118. Proc Natl Acad Sci U S A. 2021. PMID: 34187888 Free PMC article.
-
Comparative Analysis of Nano-Bactericides and Thiodiazole-Copper on Tomato Rhizosphere Microbiome.Microorganisms. 2025 Jun 7;13(6):1327. doi: 10.3390/microorganisms13061327. Microorganisms. 2025. PMID: 40572215 Free PMC article.
References
-
- Pei J, Cai W, Kinch LN, Grishin NV. Prediction of functional specificity determinants from protein sequences using log-likelihood ratios. Bioinformatics. 2006;22:164–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

