An updated evolutionary classification of CRISPR-Cas systems
- PMID: 26411297
- PMCID: PMC5426118
- DOI: 10.1038/nrmicro3569
An updated evolutionary classification of CRISPR-Cas systems
Abstract
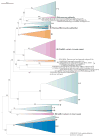
The evolution of CRISPR-cas loci, which encode adaptive immune systems in archaea and bacteria, involves rapid changes, in particular numerous rearrangements of the locus architecture and horizontal transfer of complete loci or individual modules. These dynamics complicate straightforward phylogenetic classification, but here we present an approach combining the analysis of signature protein families and features of the architecture of cas loci that unambiguously partitions most CRISPR-cas loci into distinct classes, types and subtypes. The new classification retains the overall structure of the previous version but is expanded to now encompass two classes, five types and 16 subtypes. The relative stability of the classification suggests that the most prevalent variants of CRISPR-Cas systems are already known. However, the existence of rare, currently unclassifiable variants implies that additional types and subtypes remain to be characterized.
Conflict of interest statement
The authors declare no competing interests.
Figures


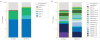
References
-
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

