Chromatin Architecture of the Pitx2 Locus Requires CTCF- and Pitx2-Dependent Asymmetry that Mirrors Embryonic Gut Laterality
- PMID: 26411685
- PMCID: PMC4617833
- DOI: 10.1016/j.celrep.2015.08.075
Chromatin Architecture of the Pitx2 Locus Requires CTCF- and Pitx2-Dependent Asymmetry that Mirrors Embryonic Gut Laterality
Abstract
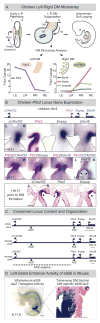
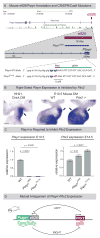
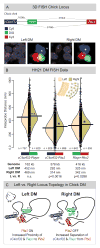
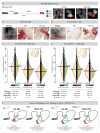
Expression of Pitx2 on the left side of the embryo patterns left-right (LR) organs including the dorsal mesentery (DM), whose asymmetric cell behavior directs gut looping. Despite the importance of organ laterality, chromatin-level regulation of Pitx2 remains undefined. Here, we show that genes immediately neighboring Pitx2 in chicken and mouse, including a long noncoding RNA (Pitx2 locus-asymmetric regulated RNA or Playrr), are expressed on the right side and repressed by Pitx2. CRISPR/Cas9 genome editing of Playrr, 3D fluorescent in situ hybridization (FISH), and variations of chromatin conformation capture (3C) demonstrate that mutual antagonism between Pitx2 and Playrr is coordinated by asymmetric chromatin interactions dependent on Pitx2 and CTCF. We demonstrate that transcriptional and morphological asymmetries driving gut looping are mirrored by chromatin architectural asymmetries at the Pitx2 locus. We propose a model whereby Pitx2 auto-regulation directs chromatin topology to coordinate LR transcription of this locus essential for LR organogenesis.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. - PubMed
-
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EEM. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

