Next-Generation AUC: Analysis of Multiwavelength Analytical Ultracentrifugation Data
- PMID: 26412646
- PMCID: PMC11583009
- DOI: 10.1016/bs.mie.2015.04.013
Next-Generation AUC: Analysis of Multiwavelength Analytical Ultracentrifugation Data
Abstract
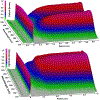
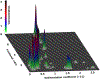
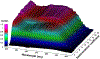
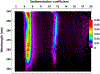
We describe important advances in methodologies for the analysis of multiwavelength data. In contrast to the Beckman-Coulter XL-A/I ultraviolet-visible light detector, multiwavelength detection is able to simultaneously collect sedimentation data for a large wavelength range in a single experiment. The additional dimension increases the data density by orders of magnitude, posing new challenges for data analysis and management. The additional data not only improve the statistics of the measurement but also provide new information for spectral characterization, which complements the hydrodynamic information. New data analysis and management approaches were integrated into the UltraScan software to address these challenges. In this chapter, we describe the enhancements and benefits realized by multiwavelength analysis and compare the results to those obtained from the traditional single-wavelength detector. We illustrate the advances offered by the new instruments by comparing results from mixtures that contain different ratios of protein and DNA samples, representing analytes with distinct spectral and hydrodynamic properties. For the first time, we demonstrate that the spectral dimension not only adds valuable detail, but when spectral properties are known, individual components with distinct spectral properties measured in a mixture by the multiwavelength system can be clearly separated and decomposed into traditional datasets for each of the spectrally distinct components, even when their sedimentation coefficients are virtually identical.
Keywords: Analytical ultracentrifugation; Hydrodynamic analysis; Multiwavelength detector; Optical detector development; Spectral analysis; UV–Vis absorbance.
© 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Bhattacharyya SK, Maciejewska P, Börger L, Stadler M, Gülsün AM, Cicek HB, et al. (2006). Development of fast fiber based UV-Vis multiwavelength detector for an ultracentrifuge. Progress in Colloid and Polymer Science, 131, 9–22.
-
- Brookes E, Boppana RV, & Demeler B (2006). Computing large sparse multivariate optimization problems with an application in biophysics. In Supercomputing ‘06 ACM 0-7695-2700-0/06.
-
- Brookes E, Cao W, & Demeler B (2010). A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. European Biophysics Journal, 39(3), 405–414. - PubMed
-
- Brookes E, & Demeler B (2006). Genetic algorithm optimization for obtaining accurate molecular weight distributions from sedimentation velocity experiments. In Wandrey C & Cölfen H (Eds.), Progress in Colloid and Polymer Science: 131. Analytical ultracentrifugation VIII (pp. 33–40): Berlin Heidelberg: Springer-Verlag. 10.1007/2882_004. - DOI
-
- Brookes E, & Demeler B (2007). Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments. In GECCO proceedings ACM 978-1-59593-697-4/07/0007.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

