Analytical Ultracentrifugation as a Tool to Study Nonspecific Protein-DNA Interactions
- PMID: 26412658
- PMCID: PMC5009906
- DOI: 10.1016/bs.mie.2015.04.009
Analytical Ultracentrifugation as a Tool to Study Nonspecific Protein-DNA Interactions
Abstract
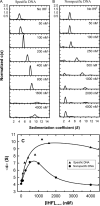
Analytical ultracentrifugation (AUC) is a powerful tool that can provide thermodynamic information on associating systems. Here, we discuss how to use the two fundamental AUC applications, sedimentation velocity (SV), and sedimentation equilibrium (SE), to study nonspecific protein-nucleic acid interactions, with a special emphasis on how to analyze the experimental data to extract thermodynamic information. We discuss three specific applications of this approach: (i) determination of nonspecific binding stoichiometry of E. coli integration host factor protein to dsDNA, (ii) characterization of nonspecific binding properties of Adenoviral IVa2 protein to dsDNA using SE-AUC, and (iii) analysis of the competition between specific and nonspecific DNA-binding interactions observed for E. coli integration host factor protein assembly on dsDNA. These approaches provide powerful tools that allow thermodynamic interrogation and thus a mechanistic understanding of how proteins bind nucleic acids by both specific and nonspecific interactions.
Keywords: Nonspecific binding; Specific and nonspecific competitive binding.
© 2015 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation.Curr Protoc Protein Sci. 2013 Feb;Chapter 20:Unit20.12. doi: 10.1002/0471140864.ps2012s71. Curr Protoc Protein Sci. 2013. PMID: 23377850 Free PMC article.
-
Modern analytical ultracentrifugation in protein science: a tutorial review.Protein Sci. 2002 Sep;11(9):2067-79. doi: 10.1110/ps.0207702. Protein Sci. 2002. PMID: 12192063 Free PMC article. Review.
-
Characterization of the non-specific DNA binding properties of the Adenoviral IVa2 protein.Biophys Chem. 2014 Sep-Oct;193-194:1-8. doi: 10.1016/j.bpc.2014.06.005. Epub 2014 Jun 28. Biophys Chem. 2014. PMID: 25038409
-
Calculations and Publication-Quality Illustrations for Analytical Ultracentrifugation Data.Methods Enzymol. 2015;562:109-33. doi: 10.1016/bs.mie.2015.05.001. Epub 2015 Jun 16. Methods Enzymol. 2015. PMID: 26412649
-
Using Lamm-Equation modeling of sedimentation velocity data to determine the kinetic and thermodynamic properties of macromolecular interactions.Methods. 2011 May;54(1):4-15. doi: 10.1016/j.ymeth.2010.12.029. Epub 2010 Dec 25. Methods. 2011. PMID: 21187153 Free PMC article. Review.
Cited by
-
Walker-A Motif Acts to Coordinate ATP Hydrolysis with Motor Output in Viral DNA Packaging.J Mol Biol. 2016 Jul 3;428(13):2709-29. doi: 10.1016/j.jmb.2016.04.029. Epub 2016 Apr 30. J Mol Biol. 2016. PMID: 27139643 Free PMC article.
-
Physical and Functional Characterization of a Viral Genome Maturation Complex.Biophys J. 2017 Apr 25;112(8):1551-1560. doi: 10.1016/j.bpj.2017.02.041. Biophys J. 2017. PMID: 28445747 Free PMC article.
-
Analytical ultracentrifuge: an ideal tool for characterization of non-coding RNAs.Eur Biophys J. 2020 Dec;49(8):809-818. doi: 10.1007/s00249-020-01470-9. Epub 2020 Oct 16. Eur Biophys J. 2020. PMID: 33067686 Review.
-
Determination of 3- and 4-chloromethcathinone interactions with plasma proteins: study involving analytical and theoretical methods.Forensic Toxicol. 2024 Jul;42(2):111-124. doi: 10.1007/s11419-023-00677-7. Epub 2023 Dec 18. Forensic Toxicol. 2024. PMID: 38108940 Free PMC article.
References
-
- Aeling KA, Opel ML, Steffen NR, Tretyachenko-Ladokhina V, Hatfield GW, Lathrop RH, et al. Indirect recognition in sequence-specific DNA binding by Escherichia coli integration host factor: The role of DNA deformation energy. The Journal of Biological Chemistry. 2006;281(51):39236–39248. - PubMed
-
- Bujalowski W. Thermodynamic and kinetic methods of analyses of protein-nucleic acid interactions. From simpler to more complex systems. Chemical Reviews. 2006;106(2):556–606. - PubMed
-
- Bujalowski W, Lohman TM, Anderson CF. On the cooperative binding of large ligands to a one-dimensional homogeneous lattice: The generalized three-state lattice model. Biopolymers. 1989;28(9):1637–1643. - PubMed
-
- Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. The Journal of Biological Chemistry. 1988;263(10):4629–4640. - PubMed
-
- Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421(6921):423–427. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

