Comprehensive comparative homeobox gene annotation in human and mouse
- PMID: 26412852
- PMCID: PMC4584094
- DOI: 10.1093/database/bav091
Comprehensive comparative homeobox gene annotation in human and mouse
Abstract
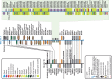
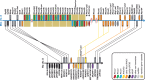
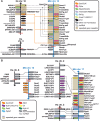
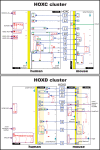
Homeobox genes are a group of genes coding for transcription factors with a DNA-binding helix-turn-helix structure called a homeodomain and which play a crucial role in pattern formation during embryogenesis. Many homeobox genes are located in clusters and some of these, most notably the HOX genes, are known to have antisense or opposite strand long non-coding RNA (lncRNA) genes that play a regulatory role. Because automated annotation of both gene clusters and non-coding genes is fraught with difficulty (over-prediction, under-prediction, inaccurate transcript structures), we set out to manually annotate all homeobox genes in the mouse and human genomes. This includes all supported splice variants, pseudogenes and both antisense and flanking lncRNAs. One of the areas where manual annotation has a significant advantage is the annotation of duplicated gene clusters. After comprehensive annotation of all homeobox genes and their antisense genes in human and in mouse, we found some discrepancies with the current gene set in RefSeq regarding exact gene structures and coding versus pseudogene locus biotype. We also identified previously un-annotated pseudogenes in the DUX, Rhox and Obox gene clusters, which helped us re-evaluate and update the gene nomenclature in these regions. We found that human homeobox genes are enriched in antisense lncRNA loci, some of which are known to play a role in gene or gene cluster regulation, compared to their mouse orthologues. Of the annotated set of 241 human protein-coding homeobox genes, 98 have an antisense locus (41%) while of the 277 orthologous mouse genes, only 62 protein coding gene have an antisense locus (22%), based on publicly available transcriptional evidence.
© The Author(s) 2015. Published by Oxford University Press.
Figures

References
-
- Qian Y.Q., Otting G., Billeter M., et al. (1993) Nuclear magnetic resonance spectroscopy of a DNA complex with the uniformly 13C-labeled Antennapedia homeodomain and structure determination of the DNA-bound homeodomain. J Mol Biol., 234, 1070–1083. - PubMed
-
- Bürglin T.R. (2013) Homeobox genes. Brenner’s Encyclopedia of Genetics. Academic Press, London.
-
- Burglin T.R. (2011) Homeodomain subtypes and functional diversity. Subcell Biochem., 52, 95–122. - PubMed
-
- Holland P.W. (2013) Evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol., 2, 31–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

