Cellulosomics of the cellulolytic thermophile Clostridium clariflavum
- PMID: 26413154
- PMCID: PMC4582956
- DOI: 10.1186/1754-6834-7-100
Cellulosomics of the cellulolytic thermophile Clostridium clariflavum
Abstract
Background: Clostridium clariflavum is an anaerobic, thermophilic, Gram-positive bacterium, capable of growth on crystalline cellulose as a single carbon source. The genome of C. clariflavum has been sequenced to completion, and numerous cellulosomal genes were identified, including putative scaffoldin and enzyme subunits.
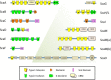
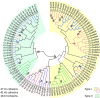
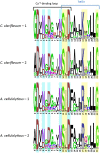
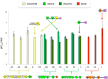
Results: Bioinformatic analysis of the C. clariflavum genome revealed 49 cohesin modules distributed on 13 different scaffoldins and 79 dockerin-containing proteins, suggesting an abundance of putative cellulosome assemblies. The 13-scaffoldin system of C. clariflavum is highly reminiscent of the proposed cellulosome system of Acetivibrio cellulolyticus. Analysis of the C. clariflavum type I dockerin sequences indicated a very high level of conservation, wherein the putative recognition residues are remarkably similar to those of A. cellulolyticus. The numerous interactions among the cellulosomal components were elucidated using a standardized affinity ELISA-based fusion-protein system. The results revealed a rather simplistic recognition pattern of cohesin-dockerin interaction, whereby the type I and type II cohesins generally recognized the dockerins of the same type. The anticipated exception to this rule was the type I dockerin of the ScaB adaptor scaffoldin which bound selectively to the type I cohesins of ScaC and ScaJ.
Conclusions: The findings reveal an intricate picture of predicted cellulosome assemblies in C. clariflavum. The network of cohesin-dockerin pairs provides a thermophilic alternative to those of C. thermocellum and a basis for subsequent utilization of the C. clariflavum cellulosomal system for biotechnological application.
Keywords: Biofuels; Biomass degradation; CBM; Cellulases; Cellulosomes; Cohesin; Dockerin; Glycoside hydrolases; Scaffoldin.
Figures


References
-
- Bayer EA, Lamed R. The cellulose paradox: pollutant par excellence and/or a reclaimable natural resource? Biodegradation. 1992;3:171–188. - PubMed
-
- Bayer EA, Lamed R, Himmel ME. The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opinion Biotechnol. 2007;18:237–245. - PubMed
-
- Atalla RH. In: Comprehensive natural products chemistry. Pinto BM, editor. Volume 3. Cambridge: Elsevier; 1999. Celluloses; pp. 529–598.
-
- O'Sullivan AC. Cellulose: the structure slowly unravels. Cellulose. 1997;4:173–207.
LinkOut - more resources
Full Text Sources
Other Literature Sources

