Insight into metabolic pathways of the potential biofuel producer, Paenibacillus polymyxa ICGEB2008
- PMID: 26413158
- PMCID: PMC4583153
- DOI: 10.1186/s13068-015-0338-4
Insight into metabolic pathways of the potential biofuel producer, Paenibacillus polymyxa ICGEB2008
Abstract
Background: Paenibacillus polymyxa is a facultative anaerobe known for production of hydrolytic enzymes and various important biofuel molecules. Despite its wide industrial use and the availability of its genome sequence, very little is known about metabolic pathways operative in the Paenibacillus system. Here, we report metabolic insights of an insect gut symbiont, Paenibacillus polymyxa ICGEB2008, and reveal pathways playing an important role in the production of 2,3-butanediol and ethanol.
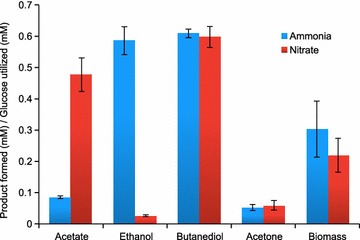
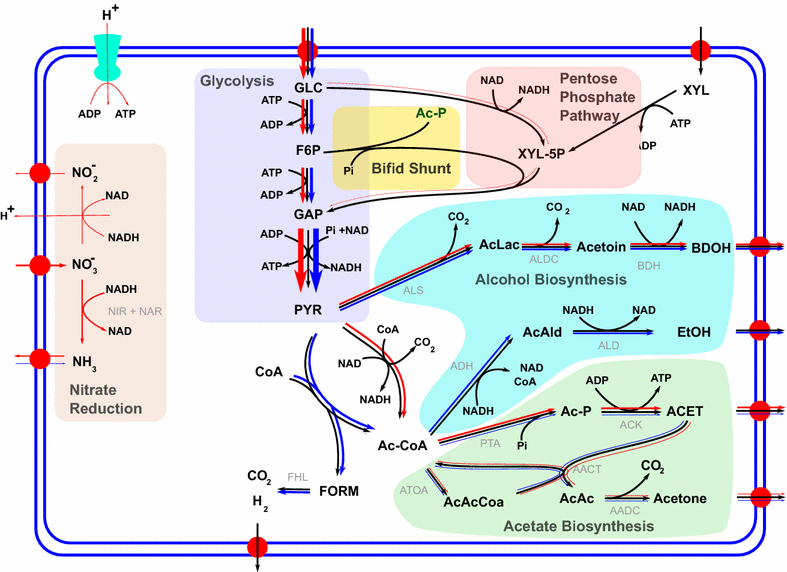
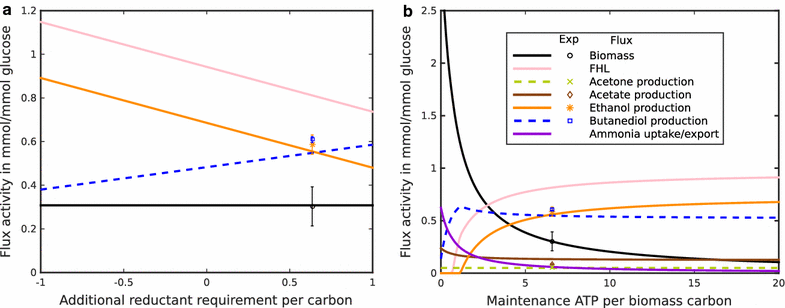
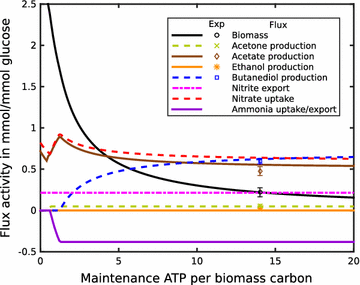
Result: We developed a metabolic network model of P. polymyxa ICGEB2008 with 133 metabolites and 158 reactions. Flux balance analysis was employed to investigate the importance of redox balance in ICGEB2008. This led to the detection of the Bifid shunt, a pathway previously not described in Paenibacillus, which can uncouple the production of ATP from the generation of reducing equivalents. Using a combined experimental and modeling approach, we further studied pathways involved in 2,3-butanediol and ethanol production and also demonstrated the production of hydrogen by the organism. We could further show that the nitrogen source is critical for metabolite production by Paenibacillus, and correctly quantify the influence on the by-product metabolite profile of ICGEB2008. Both simulations and experiments showed that metabolic flux is diverted from ethanol to acetate production when an oxidized nitrogen source is utilized.
Conclusion: We have created a predictive model of the central carbon metabolism of P. polymyxa ICGEB2008 and could show the presence of the Bifid shunt and explain its role in ICGEB2008. An in-depth study has been performed to understand the metabolic pathways involved in ethanol, 2,3-butanediol and hydrogen production, which can be utilized as a basis for further metabolic engineering efforts to improve the efficiency of biofuel production by this P. polymyxa strain.
Keywords: 2,3-Butanediol; Ethanol; Metabolic engineering; Metabolic modeling; Paenibacillus polymyxa; Redox metabolism.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

