V1/V2 Neutralizing Epitope is Conserved in Divergent Non-M Groups of HIV-1
- PMID: 26413851
- PMCID: PMC4770367
- DOI: 10.1097/QAI.0000000000000854
V1/V2 Neutralizing Epitope is Conserved in Divergent Non-M Groups of HIV-1
Abstract
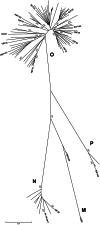
Background: Highly potent broadly neutralizing monoclonal antibodies (bNAbs) have been obtained from individuals infected by HIV-1 group M variants. We analyzed the cross-group neutralization potency of these bNAbs toward non-M primary isolates (PI).
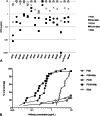
Material and methods: The sensitivity to neutralization was analyzed in a neutralization assay using TZM-bl cells. Twenty-three bNAbs were used, including reagents targeting the CD4-binding site, the N160 glycan-V1/V2 site, the N332 glycan-V3 site, the membrane proximal external region of gp41, and complex epitopes spanning both env subunits. Two bispecific antibodies that combine the inhibitory activity of an anti-CD4 with that of PG9 or PG16 bNAbs were included in the study (PG9-iMab and PG16-iMab).
Results: Cross-group neutralization was observed only with the bNAbs targeting the N160 glycan-V1/V2 site. Four group O PIs, 1 group N PI, and the group P PI were neutralized by PG9 and/or PG16 or PGT145 at low concentrations (0.04-9.39 μg/mL). None of the non-M PIs was neutralized by the bNAbs targeting other regions at the highest concentration tested, except 10E8 that neutralized weakly 2 group N PIs and 35O22 that neutralized 1 group O PI. The bispecific bNAbs neutralized very efficiently all the non-M PIs with IC50 below 1 μg/mL, except 2 group O strains.
Conclusion: The N160 glycan-V1/V2 site is the most conserved neutralizing site within the 4 groups of HIV-1. This makes it an interesting target for the development of HIV vaccine immunogens. The corresponding bNAbs may be useful for immunotherapeutic strategies in patients infected by non-M variants.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures

References
-
- Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2012;13:693–701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

