Clustering reveals limits of parameter identifiability in multi-parameter models of biochemical dynamics
- PMID: 26415494
- PMCID: PMC4587803
- DOI: 10.1186/s12918-015-0205-8
Clustering reveals limits of parameter identifiability in multi-parameter models of biochemical dynamics
Abstract
Background: Compared to engineering or physics problems, dynamical models in quantitative biology typically depend on a relatively large number of parameters. Progress in developing mathematics to manipulate such multi-parameter models and so enable their efficient interplay with experiments has been slow. Existing solutions are significantly limited by model size.
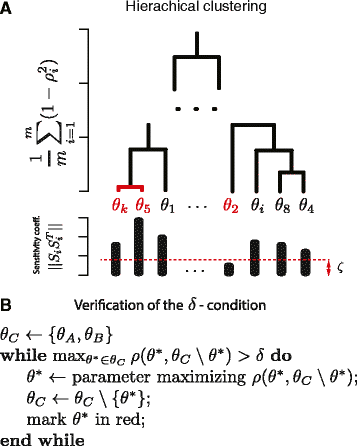
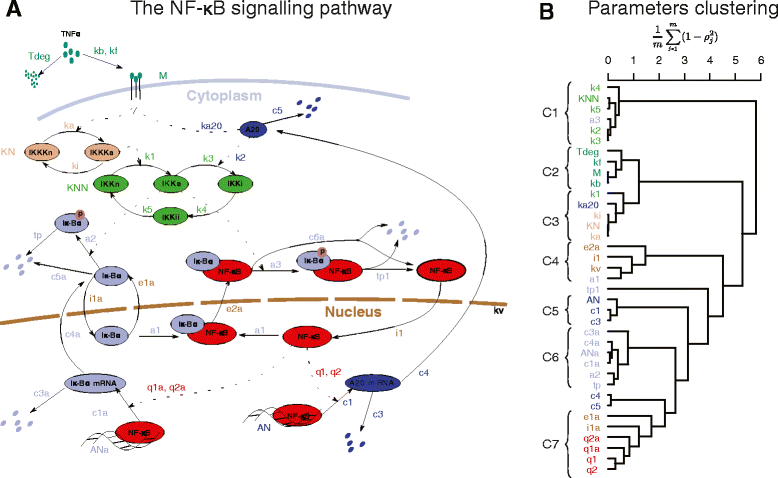
Results: In order to simplify analysis of multi-parameter models a method for clustering of model parameters is proposed. It is based on a derived statistically meaningful measure of similarity between groups of parameters. The measure quantifies to what extend changes in values of some parameters can be compensated by changes in values of other parameters. The proposed methodology provides a natural mathematical language to precisely communicate and visualise effects resulting from compensatory changes in values of parameters. As a results, a relevant insight into identifiability analysis and experimental planning can be obtained. Analysis of NF-κB and MAPK pathway models shows that highly compensative parameters constitute clusters consistent with the network topology. The method applied to examine an exceptionally rich set of published experiments on the NF-κB dynamics reveals that the experiments jointly ensure identifiability of only 60% of model parameters. The method indicates which further experiments should be performed in order to increase the number of identifiable parameters.
Conclusions: We currently lack methods that simplify broadly understood analysis of multi-parameter models. The introduced tools depict mutually compensative effects between parameters to provide insight regarding role of individual parameters, identifiability and experimental design. The method can also find applications in related methodological areas of model simplification and parameters estimation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

