Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells
- PMID: 26415882
- PMCID: PMC4587679
- DOI: 10.1186/s13059-015-0768-0
Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells
Abstract
Background: Higher-order chromatin structure is often perturbed in cancer and other pathological states. Although several genetic and epigenetic differences have been charted between normal and breast cancer tissues, changes in higher-order chromatin organization during tumorigenesis have not been fully explored. To probe the differences in higher-order chromatin structure between mammary epithelial and breast cancer cells, we performed Hi-C analysis on MCF-10A mammary epithelial and MCF-7 breast cancer cell lines.
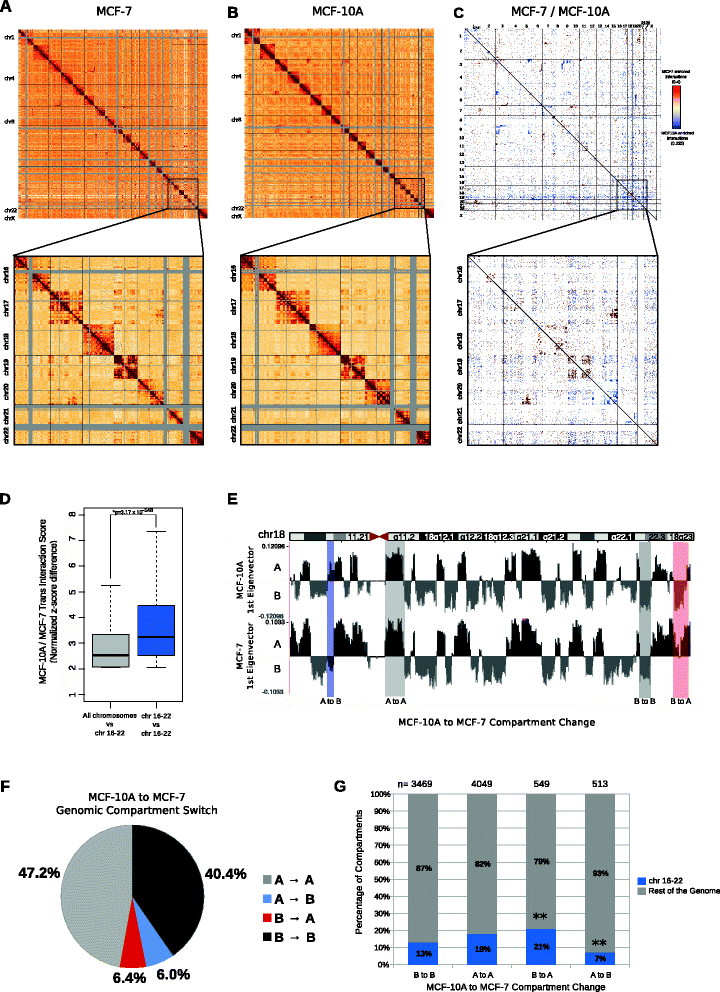
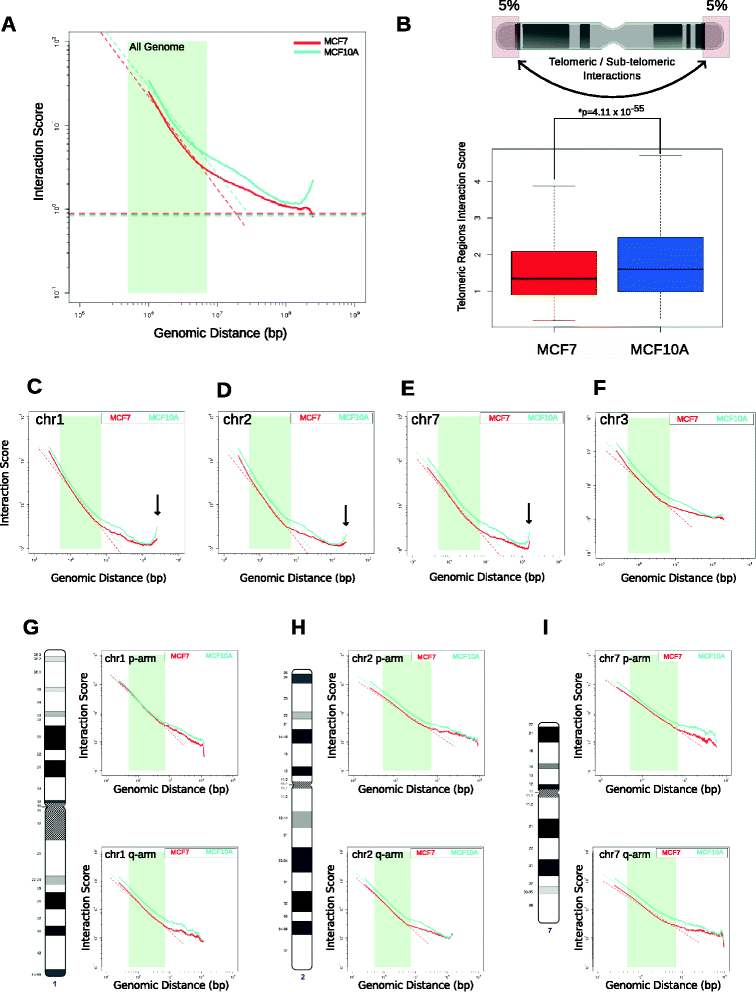
Results: Our studies reveal that the small, gene-rich chromosomes chr16 through chr22 in the MCF-7 breast cancer genome display decreased interaction frequency with each other compared to the inter-chromosomal interaction frequency in the MCF-10A epithelial cells. Interestingly, this finding is associated with a higher occurrence of open compartments on chr16-22 in MCF-7 cells. Pathway analysis of the MCF-7 up-regulated genes located in altered compartment regions on chr16-22 reveals pathways related to repression of WNT signaling. There are also differences in intra-chromosomal interactions between the cell lines; telomeric and sub-telomeric regions in the MCF-10A cells display more frequent interactions than are observed in the MCF-7 cells.
Conclusions: We show evidence of an intricate relationship between chromosomal organization and gene expression between epithelial and breast cancer cells. Importantly, this work provides a genome-wide view of higher-order chromatin dynamics and a resource for studying higher-order chromatin interactions in two cell lines commonly used to study the progression of breast cancer.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

