Important contribution of the novel locus comEB to extracellular DNA-dependent Staphylococcus lugdunensis biofilm formation
- PMID: 26416910
- PMCID: PMC4645410
- DOI: 10.1128/IAI.00775-15
Important contribution of the novel locus comEB to extracellular DNA-dependent Staphylococcus lugdunensis biofilm formation
Abstract
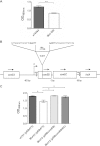
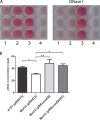
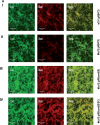
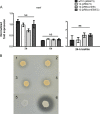
The coagulase-negative species Staphylococcus lugdunensis is an emerging cause of serious and potentially life-threatening infections, such as infective endocarditis. The pathogenesis of these infections is characterized by the ability of S. lugdunensis to form biofilms on either biotic or abiotic surfaces. To elucidate the genetic basis of biofilm formation in S. lugdunensis, we performed transposon (Tn917) mutagenesis. One mutant had a significantly reduced biofilm-forming capacity and carried a Tn917 insertion within the competence gene comEB. Site-directed mutagenesis and subsequent complementation with a functional copy of comEB verified the importance of comEB in biofilm formation. In several bacterial species, natural competence stimulates DNA release via lysis-dependent or -independent mechanisms. Extracellular DNA (eDNA) has been demonstrated to be an important structural component of many bacterial biofilms. Therefore, we quantified the eDNA in the biofilms and found diminished eDNA amounts in the comEB mutant biofilm. High-resolution images and three-dimensional data obtained via confocal laser scanning microscopy (CSLM) visualized the impact of the comEB mutation on biofilm integrity. The comEB mutant did not show reduced expression of autolysin genes, decreased autolytic activities, or increased cell viability, suggesting a cell lysis-independent mechanism of DNA release. Furthermore, reduced amounts of eDNA in the comEB mutant biofilms did not result from elevated levels or activity of the S. lugdunensis thermonuclease NucI. In conclusion, we defined here, for the first time, a role for the competence gene comEB in staphylococcal biofilm formation. Our findings indicate that comEB stimulates biofilm formation via a lysis-independent mechanism of DNA release.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Freney J, Brun Y, Bes M, Meugnier H, Grimont F, Grimont PAD, Nervi C, Fleurette J. 1988. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int J Syst Bacteriol 38:168–172. doi:10.1099/00207713-38-2-168. - DOI
-
- Ravaioli S, Selan L, Visai L, Pirini V, Campoccia D, Maso A, Speziale P, Montanaro L, Arciola CR. 2012. Staphylococcus lugdunensis, an aggressive coagulase-negative pathogen not to be underestimated. Int J Artif Organs 35:742–753. - PubMed
-
- Anguera I, Del Rio A, Miro JM, Matinez-Lacasa X, Marco F, Guma JR, Quaglio G, Claramonte X, Moreno A, Mestres CA, Mauri E, Azqueta M, Benito N, Garcia-de la Maria C, Almela M, Jimenez-Exposito MJ, Sued O, De Lazzari E, Gatell JM. 2005. Staphylococcus lugdunensis infective endocarditis: description of 10 cases and analysis of native valve, prosthetic valve, and pacemaker lead endocarditis clinical profiles. Heart 91:e10. doi:10.1136/hrt.2004.040659. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

