PDK2-mediated alternative splicing switches Bnip3 from cell death to cell survival
- PMID: 26416963
- PMCID: PMC4586742
- DOI: 10.1083/jcb.201504047
PDK2-mediated alternative splicing switches Bnip3 from cell death to cell survival
Abstract
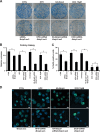
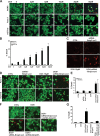
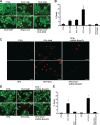
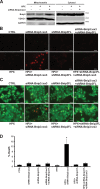
Herein we describe a novel survival pathway that operationally links alternative pre-mRNA splicing of the hypoxia-inducible death protein Bcl-2 19-kD interacting protein 3 (Bnip3) to the unique glycolytic phenotype in cancer cells. While a full-length Bnip3 protein (Bnip3FL) encoded by exons 1-6 was expressed as an isoform in normal cells and promoted cell death, a truncated spliced variant of Bnip3 mRNA deleted for exon 3 (Bnip3Δex3) was preferentially expressed in several human adenocarcinomas and promoted survival. Reciprocal inhibition of the Bnip3Δex3/Bnip3FL isoform ratio by inhibiting pyruvate dehydrogenase kinase isoform 2 (PDK2) in Panc-1 cells rapidly induced mitochondrial perturbations and cell death. The findings of the present study reveal a novel survival pathway that functionally couples the unique glycolytic phenotype in cancer cells to hypoxia resistance via a PDK2-dependent mechanism that switches Bnip3 from cell death to survival. Discovery of the survival Bnip3Δex3 isoform may fundamentally explain how certain cells resist Bnip3 and avert death during hypoxia.
© 2015 Gang et al.
Figures
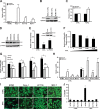
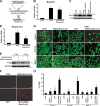
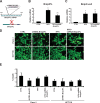
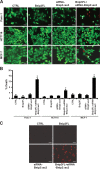
References
-
- Bonnet S., Archer S.L., Allalunis-Turner J., Haromy A., Beaulieu C., Thompson R., Lee C.T., Lopaschuk G.D., Puttagunta L., Bonnet S., et al. 2007. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 11:37–51. 10.1016/j.ccr.2006.10.020 - DOI - PubMed
-
- Chiche J., Rouleau M., Gounon P., Brahimi-Horn M.C., Pouysségur J., and Mazure N.M.. 2010. Hypoxic enlarged mitochondria protect cancer cells from apoptotic stimuli. J. Cell. Physiol. 222:648–657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

