The Roles of Compensatory Evolution and Constraint in Aminoacyl tRNA Synthetase Evolution
- PMID: 26416980
- PMCID: PMC4693975
- DOI: 10.1093/molbev/msv206
The Roles of Compensatory Evolution and Constraint in Aminoacyl tRNA Synthetase Evolution
Abstract
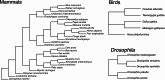
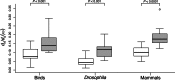
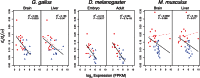
Mitochondrial protein translation requires interactions between transfer RNAs encoded by the mitochondrial genome (mt-tRNAs) and mitochondrial aminoacyl tRNA synthetase proteins (mt-aaRS) encoded by the nuclear genome. It has been argued that animal mt-tRNAs have higher deleterious substitution rates relative to their nuclear-encoded counterparts, the cytoplasmic tRNAs (cyt-tRNAs). This dynamic predicts elevated rates of compensatory evolution of mt-aaRS that interact with mt-tRNAs, relative to aaRS that interact with cyt-tRNAs (cyt-aaRS). We find that mt-aaRS do evolve at significantly higher rates (exemplified by higher dN and dN/dS) relative to cyt-aaRS, across mammals, birds, and Drosophila. While this pattern supports a model of compensatory evolution, the level at which a gene is expressed is a more general predictor of protein evolutionary rate. We find that gene expression level explains 10-56% of the variance in aaRS dN/dS, and that cyt-aaRS are more highly expressed in addition to having lower dN/dS values relative to mt-aaRS, consistent with more highly expressed genes being more evolutionarily constrained. Furthermore, we find no evidence of positive selection acting on either class of aaRS protein, as would be expected under a model of compensatory evolution. Nevertheless, the signature of faster mt-aaRS evolution persists in mammalian, but not bird or Drosophila, lineages after controlling for gene expression, suggesting some additional effect of compensatory evolution for mammalian mt-aaRS. We conclude that gene expression is the strongest factor governing differential amino acid substitution rates in proteins interacting with mitochondrial versus cytoplasmic factors, with important differences in mt-aaRS molecular evolution among taxonomic groups.
Keywords: compensatory evolution; gene expression; mitochondrial-nuclear coevolution; protein evolution.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Anisimova M, Bielawski JP, Yang Z. 2002. Accuracy and power of bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 19:950–958. - PubMed
-
- Ballard J. 2000. Comparative genomics of mitochondrial DNA in Drosophila simulans. J Mol Evol. 51:64–75. - PubMed
-
- Barreto FS, Burton RS. 2013. Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol Biol Evol. 30:310–314. - PubMed
-
- Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. 2007. The delayed rise of present-day mammals. Nature 446:507–512. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

