Floral Induction in Arabidopsis by FLOWERING LOCUS T Requires Direct Repression of BLADE-ON-PETIOLE Genes by the Homeodomain Protein PENNYWISE
- PMID: 26417007
- PMCID: PMC4634070
- DOI: 10.1104/pp.15.00960
Floral Induction in Arabidopsis by FLOWERING LOCUS T Requires Direct Repression of BLADE-ON-PETIOLE Genes by the Homeodomain Protein PENNYWISE
Abstract
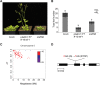
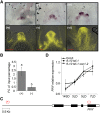
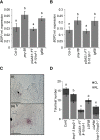
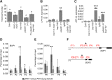
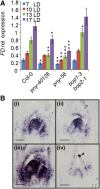
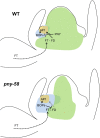
Flowers form on the flanks of the shoot apical meristem (SAM) in response to environmental and endogenous cues. In Arabidopsis (Arabidopsis thaliana), the photoperiodic pathway acts through FLOWERING LOCUS T (FT) to promote floral induction in response to day length. A complex between FT and the basic leucine-zipper transcription factor FD is proposed to form in the SAM, leading to activation of APETALA1 and LEAFY and thereby promoting floral meristem identity. We identified mutations that suppress FT function and recovered a new allele of the homeodomain transcription factor PENNYWISE (PNY). Genetic and molecular analyses showed that ectopic expression of BLADE-ON-PETIOLE1 (BOP1) and BOP2, which encode transcriptional coactivators, in the SAM during vegetative development, confers the late flowering of pny mutants. In wild-type plants, BOP1 and BOP2 are expressed in lateral organs close to boundaries of the SAM, whereas in pny mutants, their expression occurs in the SAM. This ectopic expression lowers FD mRNA levels, reducing responsiveness to FT and impairing activation of APETALA1 and LEAFY. We show that PNY binds to the promoters of BOP1 and BOP2, repressing their transcription. These results demonstrate a direct role for PNY in defining the spatial expression patterns of boundary genes and the significance of this process for floral induction by FT.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures
References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 - PubMed
-
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 - PubMed
-
- Andrés F, Porri A, Torti S, Mateos J, Romera-Branchat M, García-Martínez JL, Fornara F, Gregis V, Kater MM, Coupland G (2014) SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA 111: E2760–E2769 - PMC - PubMed
-
- Arabidopsis Genome I; Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

