New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors
- PMID: 26417104
- PMCID: PMC4611620
- DOI: 10.1073/pnas.1512995112
New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors
Abstract
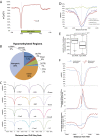
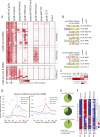
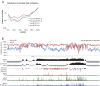
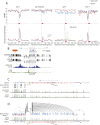
B-cell fate is orchestrated by a series of well-characterized developmental regulators. Here, we found that the onset of B-cell development was accompanied by large-scale changes in DNA cytosine modifications associated with promoters, enhancers, and anchors. These changes were tightly linked to alterations in transcription factor occupancy and nascent RNA (eRNA) transcription. We found that the prepro-B to the pro-B-cell transition was associated with a global exchange of DNA cytosine modifications for polycomb-mediated repression at CpG islands. Hypomethylated regions were found exclusively in the active/permissive compartment of the nucleus and were predominantly associated with regulatory elements or anchors that orchestrate the folding patterns of the genome. We identified superanchors, characterized by clusters of hypomethylated CCCTC-binding factor (CTCF)-bound elements, which were predominantly located at boundaries that define topological associated domains. A particularly prominent hypomethylated superanchor was positioned down-stream of the Ig heavy chain (Igh) locus. Analysis of global formaldehyde-cross-linking studies indicated that the Igh locus superanchor interacts with the VH region repertoire across vast genomic distances. We propose that the Igh locus superanchor sequesters the VH and DHJH regions into a spatial confined geometric environment to promote rapid first-passage times. Collectively, these studies demonstrate how, in developing B cells, DNA cytosine modifications associated with regulatory and architectural elements affect patterns of gene expression, folding patterns of the genome, and antigen receptor assembly.
Keywords: DNA modification; immunoglobulin heavy chain locus; nuclear architecture; superanchor; superinsulator.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. - PubMed
-
- Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26(6):715–725. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

