A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases
- PMID: 26419432
- PMCID: PMC4588251
- DOI: 10.1186/s13073-015-0221-8
A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases
Abstract
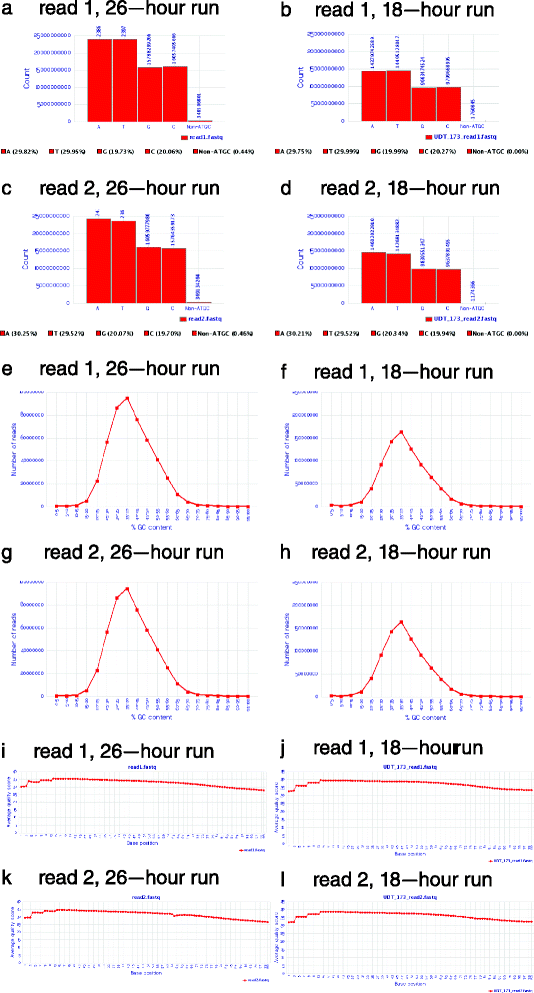
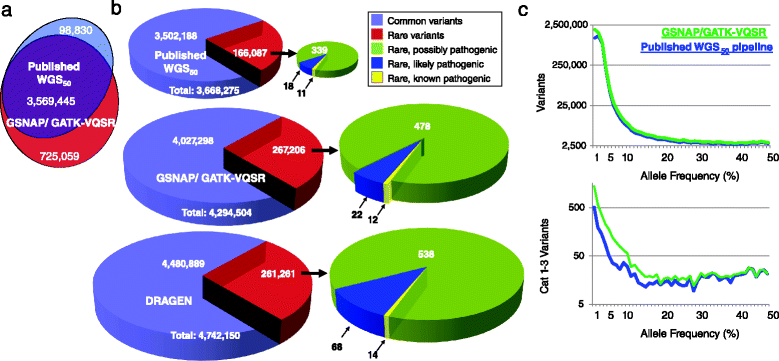
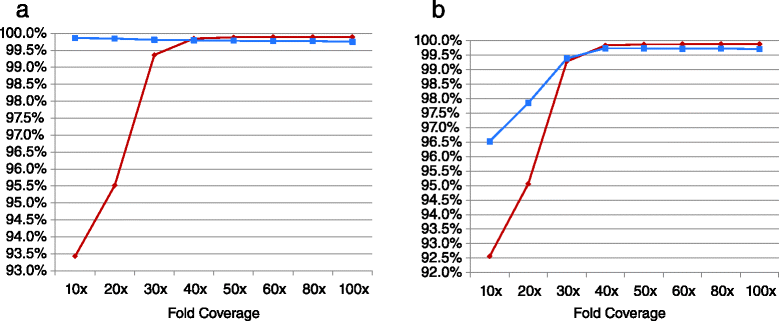
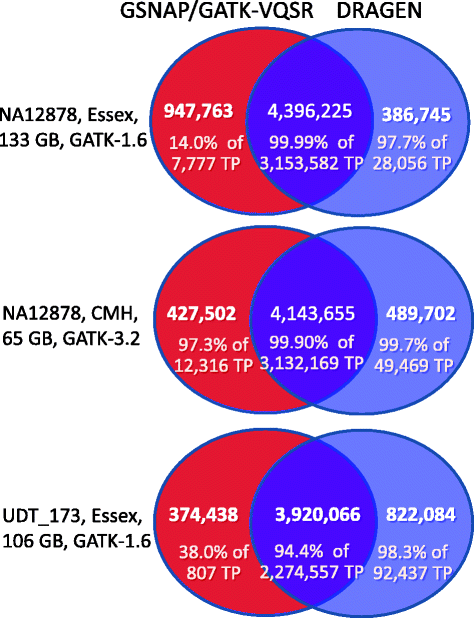
While the cost of whole genome sequencing (WGS) is approaching the realm of routine medical tests, it remains too tardy to help guide the management of many acute medical conditions. Rapid WGS is imperative in light of growing evidence of its utility in acute care, such as in diagnosis of genetic diseases in very ill infants, and genotype-guided choice of chemotherapy at cancer relapse. In such situations, delayed, empiric, or phenotype-based clinical decisions may meet with substantial morbidity or mortality. We previously described a rapid WGS method, STATseq, with a sensitivity of >96 % for nucleotide variants that allowed a provisional diagnosis of a genetic disease in 50 h. Here improvements in sequencing run time, read alignment, and variant calling are described that enable 26-h time to provisional molecular diagnosis with >99.5 % sensitivity and specificity of genotypes. STATseq appears to be an appropriate strategy for acutely ill patients with potentially actionable genetic diseases.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

