Host specificity in Sporisorium reilianum is determined by distinct mechanisms in maize and sorghum
- PMID: 26419898
- PMCID: PMC6638427
- DOI: 10.1111/mpp.12326
Host specificity in Sporisorium reilianum is determined by distinct mechanisms in maize and sorghum
Abstract
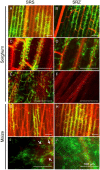
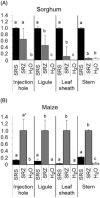
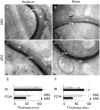
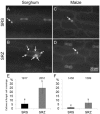
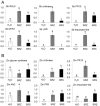
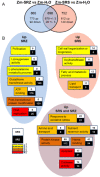
Smut fungi are biotrophic plant pathogens that exhibit a very narrow host range. The smut fungus Sporisorium reilianum exists in two host-adapted formae speciales: S. reilianum f. sp. reilianum (SRS), which causes head smut of sorghum, and S. reilianum f. sp. zeae (SRZ), which induces disease on maize. It is unknown why the two formae speciales cannot form spores on their respective non-favoured hosts. By fungal DNA quantification and fluorescence microscopy of stained plant samples, we followed the colonization behaviour of both SRS and SRZ on sorghum and maize. Both formae speciales were able to penetrate and multiply in the leaves of both hosts. In sorghum, the hyphae of SRS reached the apical meristems, whereas the hyphae of SRZ did not. SRZ strongly induced several defence responses in sorghum, such as the generation of H2 O2 , callose and phytoalexins, whereas the hyphae of SRS did not. In maize, both SRS and SRZ were able to spread through the plant to the apical meristem. Transcriptome analysis of colonized maize leaves revealed more genes induced by SRZ than by SRS, with many of them being involved in defence responses. Amongst the maize genes specifically induced by SRS were 11 pentatricopeptide repeat proteins. Together with the microscopic analysis, these data indicate that SRZ succumbs to plant defence after sorghum penetration, whereas SRS proliferates in a relatively undisturbed manner, but non-efficiently, on maize. This shows that host specificity is determined by distinct mechanisms in sorghum and maize.
Keywords: RNA sequencing; Sporisorium reilianum; defence responses; fluorescence microscopy; host specificity; phytopathogenic fungi; transcriptome.
© 2015 BSPP AND JOHN WILEY & SONS LTD.
Figures


References
-
- Ahuja, I. , Kissen, R. and Bones, A.M. (2012) Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 73–90. - PubMed
-
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- De Jonge, R. , van Esse, H.P. , Kombrink, A. , Shinya, T. , Desaki, Y. , Bours, R. , van der Krol, S. , Shibuya, N. , Joosten, M.H.A.J. and Thomma, B.P.H.J. (2010) Conserved fungal LysM effector Ecp6 prevents chitin‐triggered immunity in plants. Science, 329, 953–955. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

