Targeting Polycomb systems to regulate gene expression: modifications to a complex story
- PMID: 26420232
- PMCID: PMC5469428
- DOI: 10.1038/nrm4067
Targeting Polycomb systems to regulate gene expression: modifications to a complex story
Abstract
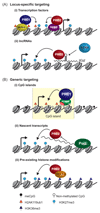
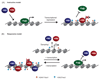
Polycomb group proteins are transcriptional repressors that are essential for normal gene regulation during development. Recent studies suggest that Polycomb repressive complexes (PRCs) recognize and are recruited to their genomic target sites through a range of different mechanisms, which involve transcription factors, CpG island elements and non-coding RNAs. Together with the realization that the interplay between PRC1 and PRC2 is more intricate than was previously appreciated, this has increased our understanding of the vertebrate Polycomb system at the molecular level.
Figures

References
-
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. - PubMed
-
- Scelfo A, Piunti A, Pasini D. The controversial role of the Polycomb group proteins in transcription and cancer: how much do we not understand Polycomb proteins? FEBS J. 2014 - PubMed
-
- Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15:340–56. - PubMed
-
- Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14:853–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

