Quantitative GSL-glycome analysis of human whole serum based on an EGCase digestion and glycoblotting method
- PMID: 26420879
- PMCID: PMC4655979
- DOI: 10.1194/jlr.D062083
Quantitative GSL-glycome analysis of human whole serum based on an EGCase digestion and glycoblotting method
Abstract
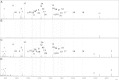
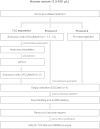
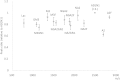
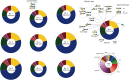
Glycosphingolipids (GSLs) are lipid molecules linked to carbohydrate units that form the plasma membrane lipid raft, which is clustered with sphingolipids, sterols, and specific proteins, and thereby contributes to membrane physical properties and specific recognition sites for various biological events. These bioactive GSL molecules consequently affect the pathophysiology and pathogenesis of various diseases. Thus, altered expression of GSLs in various diseases may be of importance for disease-related biomarker discovery. However, analysis of GSLs in blood is particularly challenging because GSLs are present at extremely low concentrations in serum/plasma. In this study, we established absolute GSL-glycan analysis of human serum based on endoglycoceramidase digestion and glycoblotting purification. We established two sample preparation protocols, one with and the other without GSL extraction using chloroform/methanol. Similar amounts of GSL-glycans were recovered with the two protocols. Both protocols permitted absolute quantitation of GSL-glycans using as little as 20 μl of serum. Using 10 healthy human serum samples, up to 42 signals corresponding to GSL-glycan compositions could be quantitatively detected, and the total serum GSL-glycan concentration was calculated to be 12.1-21.4 μM. We further applied this method to TLC-prefractionated serum samples. These findings will assist the discovery of disease-related biomarkers by serum GSL-glycomics.
Keywords: biomarker; blood; diagnostic tools; endoglycoceramidase; gangliosides; glycolipids; glycomics; glycosphingolipid; lipidomics; mass spectrometry; thin-layer chromatography.
Copyright © 2015 by the American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Lingwood D., and Simons K.. 2010. Lipid rafts as a membrane-organizing principle. Science. 327: 46–50. - PubMed
-
- Platt F. M. 2014. Sphingolipid lysosomal storage disorders. Nature. 510: 68–75. - PubMed
-
- Ryland L. K., Fox T. E., Liu X., Loughran T. P., and Kester M.. 2011. Dysregulation of sphingolipid metabolism in cancer. Cancer Biol. Ther. 11: 138–149. - PubMed
-
- Prokazova N. V., and Bergelson L. D.. 1994. Gangliosides and atherosclerosis. Lipids. 29: 1–5. - PubMed
-
- Inokuchi J. 2014. GM3 and diabetes. Glycoconj. J. 31: 193–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

