High-coverage sequencing and annotated assembly of the genome of the Australian dragon lizard Pogona vitticeps
- PMID: 26421146
- PMCID: PMC4585809
- DOI: 10.1186/s13742-015-0085-2
High-coverage sequencing and annotated assembly of the genome of the Australian dragon lizard Pogona vitticeps
Abstract
Background: The lizards of the family Agamidae are one of the most prominent elements of the Australian reptile fauna. Here, we present a genomic resource built on the basis of a wild-caught male ZZ central bearded dragon Pogona vitticeps.
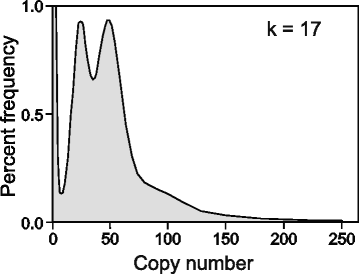
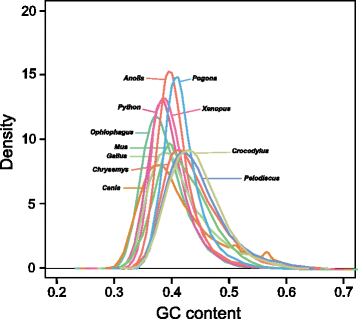
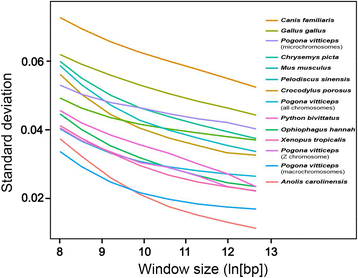
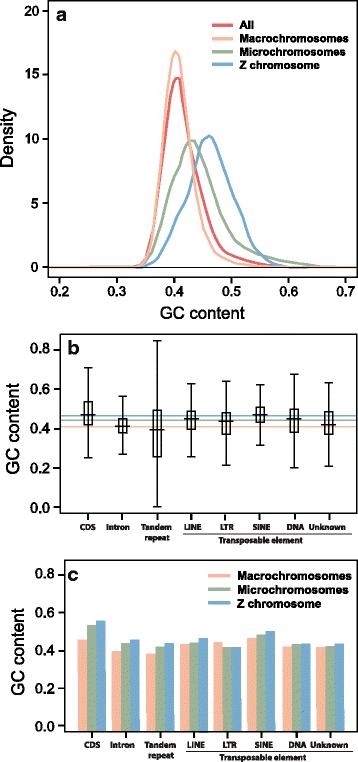
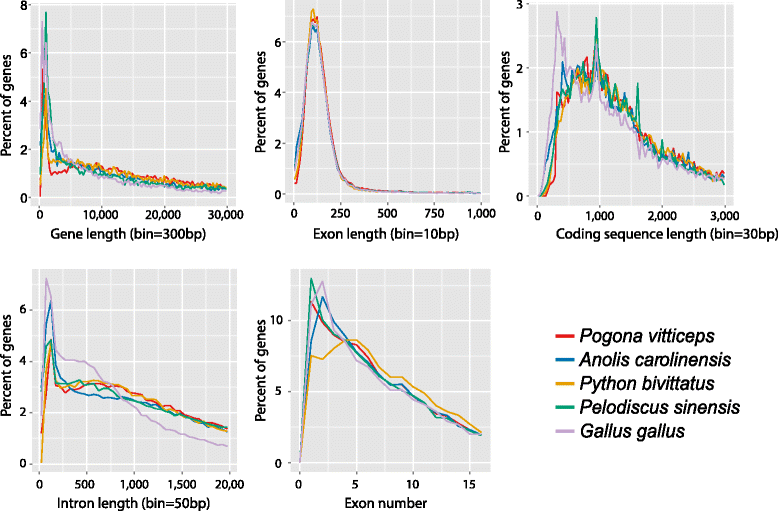
Findings: The genomic sequence for P. vitticeps, generated on the Illumina HiSeq 2000 platform, comprised 317 Gbp (179X raw read depth) from 13 insert libraries ranging from 250 bp to 40 kbp. After filtering for low-quality and duplicated reads, 146 Gbp of data (83X) was available for assembly. Exceptionally high levels of heterozygosity (0.85 % of single nucleotide polymorphisms plus sequence insertions or deletions) complicated assembly; nevertheless, 96.4 % of reads mapped back to the assembled scaffolds, indicating that the assembly included most of the sequenced genome. Length of the assembly was 1.8 Gbp in 545,310 scaffolds (69,852 longer than 300 bp), the longest being 14.68 Mbp. N50 was 2.29 Mbp. Genes were annotated on the basis of de novo prediction, similarity to the green anole Anolis carolinensis, Gallus gallus and Homo sapiens proteins, and P. vitticeps transcriptome sequence assemblies, to yield 19,406 protein-coding genes in the assembly, 63 % of which had intact open reading frames. Our assembly captured 99 % (246 of 248) of core CEGMA genes, with 93 % (231) being complete.
Conclusions: The quality of the P. vitticeps assembly is comparable or superior to that of other published squamate genomes, and the annotated P. vitticeps genome can be accessed through a genome browser available at https://genomics.canberra.edu.au.
Keywords: Agamidae, Squamata, Next-generation sequencing; Central bearded dragon; Dragon lizard; Pogona vitticeps.
Figures
Similar articles
-
Anchoring genome sequence to chromosomes of the central bearded dragon (Pogona vitticeps) enables reconstruction of ancestral squamate macrochromosomes and identifies sequence content of the Z chromosome.BMC Genomics. 2016 Jun 10;17:447. doi: 10.1186/s12864-016-2774-3. BMC Genomics. 2016. PMID: 27286959 Free PMC article.
-
A near telomere-to-telomere phased genome assembly and annotation for the Australian central bearded dragon Pogona vitticeps.Gigascience. 2025 Jan 6;14:giaf085. doi: 10.1093/gigascience/giaf085. Gigascience. 2025. PMID: 40825569 Free PMC article.
-
Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae).Chromosome Res. 2013 Jul;21(4):361-74. doi: 10.1007/s10577-013-9362-z. Epub 2013 May 24. Chromosome Res. 2013. PMID: 23703235
-
Diagnostic Clinical Pathology of the Bearded Dragon (Pogona vitticeps).Vet Clin North Am Exot Anim Pract. 2022 Sep;25(3):713-734. doi: 10.1016/j.cvex.2022.06.002. Vet Clin North Am Exot Anim Pract. 2022. PMID: 36122948 Review.
-
A lizard is never late: Squamate genomics as a recent catalyst for understanding sex chromosome and microchromosome evolution.J Hered. 2023 Aug 23;114(5):445-458. doi: 10.1093/jhered/esad023. J Hered. 2023. PMID: 37018459 Free PMC article.
Cited by
-
Immune and sex-biased gene expression in the threatened Mojave desert tortoise, Gopherus agassizii.PLoS One. 2020 Aug 26;15(8):e0238202. doi: 10.1371/journal.pone.0238202. eCollection 2020. PLoS One. 2020. PMID: 32846428 Free PMC article.
-
Spatial tissue profiling by imaging-free molecular tomography.Nat Biotechnol. 2021 Aug;39(8):968-977. doi: 10.1038/s41587-021-00879-7. Epub 2021 Apr 19. Nat Biotechnol. 2021. PMID: 33875865
-
Sex Chromosomes and Master Sex-Determining Genes in Turtles and Other Reptiles.Genes (Basel). 2021 Nov 19;12(11):1822. doi: 10.3390/genes12111822. Genes (Basel). 2021. PMID: 34828428 Free PMC article. Review.
-
Discovery of a New TLR Gene and Gene Expansion Event through Improved Desert Tortoise Genome Assembly with Chromosome-Scale Scaffolds.Genome Biol Evol. 2020 Feb 1;12(2):3917-3925. doi: 10.1093/gbe/evaa016. Genome Biol Evol. 2020. PMID: 32011707 Free PMC article.
-
The developmental origins of heterodonty and acrodonty as revealed by reptile dentitions.Sci Adv. 2021 Dec 17;7(51):eabj7912. doi: 10.1126/sciadv.abj7912. Epub 2021 Dec 17. Sci Adv. 2021. PMID: 34919438 Free PMC article.
References
-
- MacCulloch RD, Upton DE, Murphy RW. Trends in nuclear DNA content among amphibians and reptiles. Comp Biochem Physiol. 1996;113B:601–5. doi: 10.1016/0305-0491(95)02033-0. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

