Whole Genomic Analysis of an Unusual Human G6P[14] Rotavirus Strain Isolated from a Child with Diarrhea in Thailand: Evidence for Bovine-To-Human Interspecies Transmission and Reassortment Events
- PMID: 26421718
- PMCID: PMC4589232
- DOI: 10.1371/journal.pone.0139381
Whole Genomic Analysis of an Unusual Human G6P[14] Rotavirus Strain Isolated from a Child with Diarrhea in Thailand: Evidence for Bovine-To-Human Interspecies Transmission and Reassortment Events
Abstract
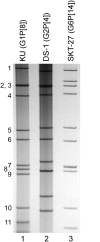
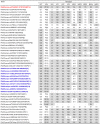
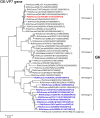
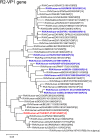
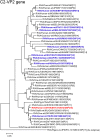
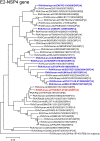
An unusual rotavirus strain, SKT-27, with the G6P[14] genotypes (RVA/Human-wt/THA/SKT-27/2012/G6P[14]), was identified in a stool specimen from a hospitalized child aged eight months with severe diarrhea. In this study, we sequenced and characterized the complete genome of strain SKT-27. On whole genomic analysis, strain SKT-27 was found to have a unique genotype constellation: G6-P[14]-I2-R2-C2-M2-A3-N2-T6-E2-H3. The non-G/P genotype constellation of this strain (I2-R2-C2-M2-A3-N2-T6-E2-H3) is commonly shared with rotavirus strains from artiodactyls such as cattle. Phylogenetic analysis indicated that nine of the 11 genes of strain SKT-27 (VP7, VP4, VP6, VP2-3, NSP1, NSP3-5) appeared to be of artiodactyl (likely bovine) origin, while the remaining VP1 and NSP2 genes were assumed to be of human origin. Thus, strain SKT-27 was found to have a bovine rotavirus genetic backbone, and thus is likely to be of bovine origin. Furthermore, strain SKT-27 appeared to be derived through interspecies transmission and reassortment events involving bovine and human rotavirus strains. Of note is that the VP7 gene of strain SKT-27 was located in G6 lineage-5 together with those of bovine rotavirus strains, away from the clusters comprising other G6P[14] strains in G6 lineages-2/6, suggesting the occurrence of independent bovine-to-human interspecies transmission events. To our knowledge, this is the first report on full genome-based characterization of human G6P[14] strains that have emerged in Southeast Asia. Our observations will provide important insights into the origin of G6P[14] strains, and into dynamic interactions between human and bovine rotavirus strains.
Conflict of interest statement
Figures







Similar articles
-
Reassortment of Human and Animal Rotavirus Gene Segments in Emerging DS-1-Like G1P[8] Rotavirus Strains.PLoS One. 2016 Feb 4;11(2):e0148416. doi: 10.1371/journal.pone.0148416. eCollection 2016. PLoS One. 2016. PMID: 26845439 Free PMC article.
-
Full Genome Characterization of Novel DS-1-Like G8P[8] Rotavirus Strains that Have Emerged in Thailand: Reassortment of Bovine and Human Rotavirus Gene Segments in Emerging DS-1-Like Intergenogroup Reassortant Strains.PLoS One. 2016 Nov 1;11(11):e0165826. doi: 10.1371/journal.pone.0165826. eCollection 2016. PLoS One. 2016. PMID: 27802339 Free PMC article.
-
Characterization of a G10P[14] rotavirus strain from a diarrheic child in Thailand: Evidence for bovine-to-human zoonotic transmission.Infect Genet Evol. 2018 Sep;63:43-57. doi: 10.1016/j.meegid.2018.05.009. Epub 2018 May 15. Infect Genet Evol. 2018. PMID: 29772399
-
[The possibilities of zoonotic transmission of rotaviruses].Epidemiol Mikrobiol Imunol. 2015 Jun;64(2):66-71. Epidemiol Mikrobiol Imunol. 2015. PMID: 26099609 Review. Czech.
-
Molecular epidemiology of human rotaviruses: genogrouping by RNA-RNA hybridization.Arch Virol Suppl. 1996;12:93-8. doi: 10.1007/978-3-7091-6553-9_11. Arch Virol Suppl. 1996. PMID: 9015106 Review.
Cited by
-
Full genome characterization of a Kenyan G8P[14] rotavirus strain suggests artiodactyl-to-human zoonotic transmission.Trop Med Health. 2025 Jun 16;53(1):82. doi: 10.1186/s41182-025-00759-9. Trop Med Health. 2025. PMID: 40524227 Free PMC article.
-
G and P genotype profiles of rotavirus a field strains circulating in a vaccinated bovine farm as parameters for assessing biosecurity level.J Vet Med Sci. 2022 Jul 1;84(7):929-937. doi: 10.1292/jvms.22-0026. Epub 2022 May 4. J Vet Med Sci. 2022. PMID: 35527015 Free PMC article.
-
Molecular characterization of the first G24P[14] rotavirus strain detected in humans.Infect Genet Evol. 2016 Sep;43:338-42. doi: 10.1016/j.meegid.2016.05.033. Epub 2016 May 26. Infect Genet Evol. 2016. PMID: 27237948 Free PMC article.
-
Evidence for zoonotic transmission of species A rotavirus from goat and cattle in nomadic herds in Morocco, 2012-2014.Virus Genes. 2020 Oct;56(5):582-593. doi: 10.1007/s11262-020-01778-w. Epub 2020 Jul 10. Virus Genes. 2020. PMID: 32651833 Free PMC article.
-
A Multiplex PCR/LDR Assay for Viral Agents of Diarrhea with the Capacity to Genotype Rotavirus.Sci Rep. 2018 Sep 4;8(1):13215. doi: 10.1038/s41598-018-30301-3. Sci Rep. 2018. PMID: 30181651 Free PMC article.
References
-
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Dugue J, Parashar UD, et al. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systemic review and meta-analysis. Lancet Infect Dis. 2012;12: 136–141. 10.1016/S1473-3099(11)70253-5 - DOI - PubMed
-
- Kahn G, Fitzwater S, Tate J, Kang G, Ganguly N, Nair G, et al. Epidemiology and prospects for prevention of rotavirus disease in India. Indian Pediatr. 2012;49: 467–474. - PubMed
-
- Estes MK, Greenberg HB. Rotaviruses In: Knipe DM, Howley PM, editors. Fields Virology. 6th ed Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1347–1401.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

