Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies
- PMID: 26423053
- PMCID: PMC4589899
- DOI: 10.1186/s13073-015-0211-x
Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies
Abstract
Background: In addition to HLA genetic incompatibility, non-HLA difference between donor and recipients of transplantation leading to allograft rejection are now becoming evident. We aimed to create a unique genome-wide platform to facilitate genomic research studies in transplant-related studies. We designed a genome-wide genotyping tool based on the most recent human genomic reference datasets, and included customization for known and potentially relevant metabolic and pharmacological loci relevant to transplantation.
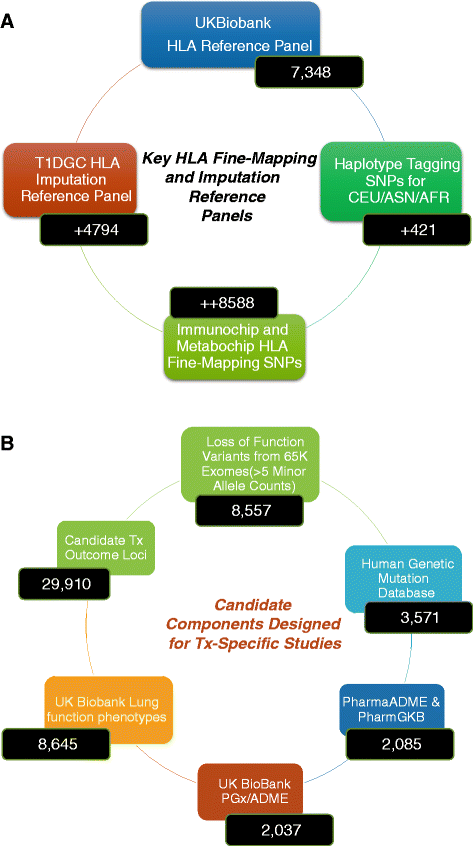
Methods: We describe here the design and implementation of a customized genome-wide genotyping array, the 'TxArray', comprising approximately 782,000 markers with tailored content for deeper capture of variants across HLA, KIR, pharmacogenomic, and metabolic loci important in transplantation. To test concordance and genotyping quality, we genotyped 85 HapMap samples on the array, including eight trios.
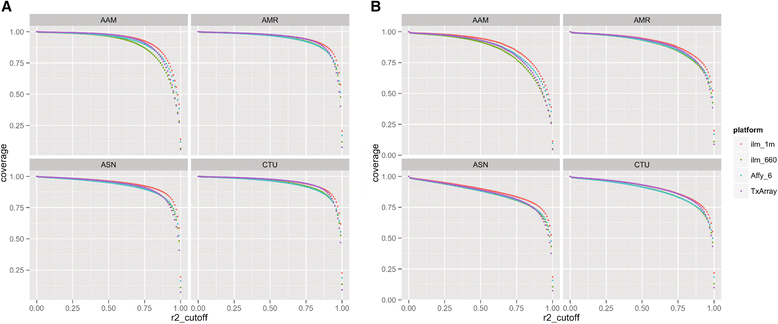
Results: We show low Mendelian error rates and high concordance rates for HapMap samples (average parent-parent-child heritability of 0.997, and concordance of 0.996). We performed genotype imputation across autosomal regions, masking directly genotyped SNPs to assess imputation accuracy and report an accuracy of >0.962 for directly genotyped SNPs. We demonstrate much higher capture of the natural killer cell immunoglobulin-like receptor (KIR) region versus comparable platforms. Overall, we show that the genotyping quality and coverage of the TxArray is very high when compared to reference samples and to other genome-wide genotyping platforms.
Conclusions: We have designed a comprehensive genome-wide genotyping tool which enables accurate association testing and imputation of ungenotyped SNPs, facilitating powerful and cost-effective large-scale genotyping of transplant-related studies.
Figures


References
-
- Organ Procurement and Transplantation Network. Available at: http://optn.transplant.hrsa.gov.
-
- Stehlik J, Edwards LB, Kucheryavaya AY, Aurora P, Christie JD, Kirk R, et al. The Registry of the International Society for Heart and Lung Transplantation: twenty-seventh official adult heart transplant report--2010. J Heart Lung Transplant. 2010;29:1089–103. doi: 10.1016/j.healun.2010.08.007. - DOI - PubMed
-
- Terasaki PI. Deduction of the fraction of immunologic and non-immunologic failure in cadaver donor transplants. Clin Transpl. 2003;449–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

