Three-dimensional matrix fiber alignment modulates cell migration and MT1-MMP utility by spatially and temporally directing protrusions
- PMID: 26423227
- PMCID: PMC4589685
- DOI: 10.1038/srep14580
Three-dimensional matrix fiber alignment modulates cell migration and MT1-MMP utility by spatially and temporally directing protrusions
Abstract
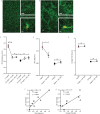
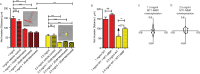
Multiple attributes of the three-dimensional (3D) extracellular matrix (ECM) have been independently implicated as regulators of cell motility, including pore size, crosslink density, structural organization, and stiffness. However, these parameters cannot be independently varied within a complex 3D ECM protein network. We present an integrated, quantitative study of these parameters across a broad range of complex matrix configurations using self-assembling 3D collagen and show how each parameter relates to the others and to cell motility. Increasing collagen density resulted in a decrease and then an increase in both pore size and fiber alignment, which both correlated significantly with cell motility but not bulk matrix stiffness within the range tested. However, using the crosslinking enzyme Transglutaminase II to alter microstructure independently of density revealed that motility is most significantly predicted by fiber alignment. Cellular protrusion rate, protrusion orientation, speed of migration, and invasion distance showed coupled biphasic responses to increasing collagen density not predicted by 2D models or by stiffness, but instead by fiber alignment. The requirement of matrix metalloproteinase (MMP) activity was also observed to depend on microstructure, and a threshold of MMP utility was identified. Our results suggest that fiber topography guides protrusions and thereby MMP activity and motility.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

