PP2A/B56 and GSK3/Ras suppress PKB activity during Dictyostelium chemotaxis
- PMID: 26424797
- PMCID: PMC4666131
- DOI: 10.1091/mbc.E14-06-1130
PP2A/B56 and GSK3/Ras suppress PKB activity during Dictyostelium chemotaxis
Abstract
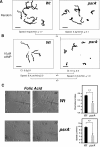
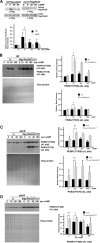
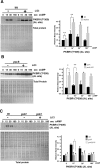
We have previously shown that the Dictyostelium protein phosphatase 2A regulatory subunit B56, encoded by psrA, modulates Dictyostelium cell differentiation through negatively affecting glycogen synthase kinase 3 (GSK3) function. Our follow-up research uncovered that B56 preferentially associated with GDP forms of RasC and RasD, but not with RasG in vitro, and psrA(-) cells displayed inefficient activation of multiple Ras species, decreased random motility, and inefficient chemotaxis toward cAMP and folic acid gradient. Surprisingly, psrA(-) cells displayed aberrantly high basal and poststimulus phosphorylation of Dictyostelium protein kinase B (PKB) kinase family member PKBR1 and PKB substrates. Expression of constitutively active Ras mutants or inhibition of GSK3 in psrA(-) cells increased activities of both PKBR1 and PKBA, but only the PKBR1 activity was increased in wild-type cells under the equivalent conditions, indicating that either B56- or GSK3-mediated suppressive mechanism is sufficient to maintain low PKBA activity, but both mechanisms are necessary for suppressing PKBR1. Finally, cells lacking RasD or RasC displayed normal PKBR1 regulation under GSK3-inhibiting conditions, indicating that RasC or RasD proteins are essential for GSK3-mediated PKBR1 inhibition. In summary, B56 constitutes inhibitory circuits for PKBA and PKBR1 and thus heavily affects Dictyostelium chemotaxis.
© 2015 Pino et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures






References
-
- Bandala-Sanchez E, Annesley SJ, Fisher PR. A phototaxis signalling complex in Dictyostelium discoideum. Eur J Cell Biol. 2006;85:1099–1106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

