Dietary Modulation of Gut Microbiota Contributes to Alleviation of Both Genetic and Simple Obesity in Children
- PMID: 26425705
- PMCID: PMC4563136
- DOI: 10.1016/j.ebiom.2015.07.007
Dietary Modulation of Gut Microbiota Contributes to Alleviation of Both Genetic and Simple Obesity in Children
Abstract
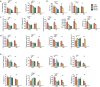
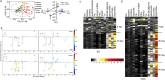
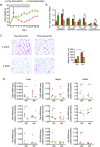
Gut microbiota has been implicated as a pivotal contributing factor in diet-related obesity; however, its role in development of disease phenotypes in human genetic obesity such as Prader-Willi syndrome (PWS) remains elusive. In this hospitalized intervention trial with PWS (n = 17) and simple obesity (n = 21) children, a diet rich in non-digestible carbohydrates induced significant weight loss and concomitant structural changes of the gut microbiota together with reduction of serum antigen load and alleviation of inflammation. Co-abundance network analysis of 161 prevalent bacterial draft genomes assembled directly from metagenomic datasets showed relative increase of functional genome groups for acetate production from carbohydrates fermentation. NMR-based metabolomic profiling of urine showed diet-induced overall changes of host metabotypes and identified significantly reduced trimethylamine N-oxide and indoxyl sulfate, host-bacteria co-metabolites known to induce metabolic deteriorations. Specific bacterial genomes that were correlated with urine levels of these detrimental co-metabolites were found to encode enzyme genes for production of their precursors by fermentation of choline or tryptophan in the gut. When transplanted into germ-free mice, the pre-intervention gut microbiota induced higher inflammation and larger adipocytes compared with the post-intervention microbiota from the same volunteer. Our multi-omics-based systems analysis indicates a significant etiological contribution of dysbiotic gut microbiota to both genetic and simple obesity in children, implicating a potentially effective target for alleviation.
Research in context: Poorly managed diet and genetic mutations are the two primary driving forces behind the devastating epidemic of obesity-related diseases. Lack of understanding of the molecular chain of causation between the driving forces and the disease endpoints retards progress in prevention and treatment of the diseases. We found that children genetically obese with Prader-Willi syndrome shared a similar dysbiosis in their gut microbiota with those having diet-related obesity. A diet rich in non-digestible but fermentable carbohydrates significantly promoted beneficial groups of bacteria and reduced toxin-producers, which contributes to the alleviation of metabolic deteriorations in obesity regardless of the primary driving forces.
Keywords: Genome interaction network; Gut microbiota; Metabolomics; Metagenomics; Obesity; Prader–Willi syndrome.
Figures



Comment in
-
Getting Healthier Through Microbiome Makeover.EBioMedicine. 2015 Aug 14;2(8):771. doi: 10.1016/j.ebiom.2015.08.024. eCollection 2015 Aug. EBioMedicine. 2015. PMID: 26425669 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

