Epigenetic Therapy for Solid Tumors: Highlighting the Impact of Tumor Hypoxia
- PMID: 26426056
- PMCID: PMC4690023
- DOI: 10.3390/genes6040935
Epigenetic Therapy for Solid Tumors: Highlighting the Impact of Tumor Hypoxia
Abstract
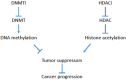
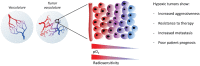
In the last few decades, epigenetics has emerged as an exciting new field in development and disease, with a more recent focus towards cancer. Epigenetics has classically referred to heritable patterns of gene expression, primarily mediated through DNA methylation patterns. More recently, it has come to include the reversible chemical modification of histones and DNA that dictate gene expression patterns. Both the epigenetic up-regulation of oncogenes and downregulation of tumor suppressors have been shown to drive tumor development. Current clinical trials for cancer therapy include pharmacological inhibition of DNA methylation and histone deacetylation, with the aim of reversing these cancer-promoting epigenetic changes. However, the DNA methyltransferase and histone deacetylase inhibitors have met with less than promising results in the treatment of solid tumors. Regions of hypoxia are a common occurrence in solid tumors. Tumor hypoxia is associated with increased aggressiveness and therapy resistance, and importantly, hypoxic tumor cells have a distinct epigenetic profile. In this review, we provide a summary of the recent clinical trials using epigenetic drugs in solid tumors, discuss the hypoxia-induced epigenetic changes and highlight the importance of testing the epigenetic drugs for efficacy against the most aggressive hypoxic fraction of the tumor in future preclinical testing.
Keywords: DNA methylation; epigenetic drugs; gene-repression; histone deacetylation; histone methylation; tumor hypoxia.
Figures
Similar articles
-
Inhibitors of DNA Methylation, Histone Deacetylation, and Histone Demethylation: A Perfect Combination for Cancer Therapy.Adv Cancer Res. 2016;130:55-111. doi: 10.1016/bs.acr.2016.01.007. Epub 2016 Mar 2. Adv Cancer Res. 2016. PMID: 27037751 Review.
-
Inhibitors of DNA Methylation and Histone Deacetylation as Epigenetically Active Drugs for Anticancer Therapy.Curr Pharm Des. 2019;25(6):635-641. doi: 10.2174/1381612825666190405144026. Curr Pharm Des. 2019. PMID: 30950345 Review.
-
3-deazaneplanocin A, a histone methyltransferase inhibitor, improved the chemoresistance induced under hypoxia in melanoma cells.Biochem Biophys Res Commun. 2023 Oct 15;677:26-30. doi: 10.1016/j.bbrc.2023.08.003. Epub 2023 Aug 2. Biochem Biophys Res Commun. 2023. PMID: 37542772
-
Hypoxia-driven epigenetic regulation in cancer progression: A focus on histone methylation and its modifying enzymes.Cancer Lett. 2020 Oct 1;489:41-49. doi: 10.1016/j.canlet.2020.05.025. Epub 2020 Jun 6. Cancer Lett. 2020. PMID: 32522693 Review.
-
Epigenetic drugs in cancer therapy.Cancer Metastasis Rev. 2025 Feb 26;44(1):37. doi: 10.1007/s10555-025-10253-7. Cancer Metastasis Rev. 2025. PMID: 40011240 Free PMC article. Review.
Cited by
-
Epigenetics in Breast Cancer Therapy-New Strategies and Future Nanomedicine Perspectives.Cancers (Basel). 2020 Dec 3;12(12):3622. doi: 10.3390/cancers12123622. Cancers (Basel). 2020. PMID: 33287297 Free PMC article. Review.
-
HIF1α-mediated transactivation of WTAP promotes AML cell proliferation via m6A-dependent stabilization of KDM4B mRNA.Leukemia. 2023 Jun;37(6):1254-1267. doi: 10.1038/s41375-023-01904-1. Epub 2023 Apr 22. Leukemia. 2023. PMID: 37087529
-
Beyond regulations at DNA levels: A review of epigenetic therapeutics targeting cancer stem cells.Cell Prolif. 2021 Feb;54(2):e12963. doi: 10.1111/cpr.12963. Epub 2020 Dec 13. Cell Prolif. 2021. PMID: 33314500 Free PMC article. Review.
-
Blood Stasis Syndrome Accelerates the Growth and Metastasis of Breast Cancer by Promoting Hypoxia and Immunosuppressive Microenvironment in Mice.J Immunol Res. 2022 Jun 7;2022:7222638. doi: 10.1155/2022/7222638. eCollection 2022. J Immunol Res. 2022. PMID: 35711625 Free PMC article.
-
DNA Strand Break Properties of Protoporphyrin IX by X-Ray Irradiation against Melanoma.Int J Mol Sci. 2020 Mar 26;21(7):2302. doi: 10.3390/ijms21072302. Int J Mol Sci. 2020. PMID: 32225109 Free PMC article.
References
-
- Tian X., Zhang S., Liu H.M., Zhang Y.B., Blair C.A., Mercola D., Sassone-Corsi P., Zi X. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: New targets for cancer therapy and prevention. Curr. Cancer Drug Targets. 2013;13:558–579. doi: 10.2174/1568009611313050007. - DOI - PMC - PubMed
-
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

