Proof-of-principle rapid noninvasive prenatal diagnosis of autosomal recessive founder mutations
- PMID: 26426075
- PMCID: PMC4607112
- DOI: 10.1172/JCI79322
Proof-of-principle rapid noninvasive prenatal diagnosis of autosomal recessive founder mutations
Abstract
Background: Noninvasive prenatal testing can be used to accurately detect chromosomal aneuploidies in circulating fetal DNA; however, the necessity of parental haplotype construction is a primary drawback to noninvasive prenatal diagnosis (NIPD) of monogenic disease. Family-specific haplotype assembly is essential for accurate diagnosis of minuscule amounts of circulating cell-free fetal DNA; however, current haplotyping techniques are too time-consuming and laborious to be carried out within the limited time constraints of prenatal testing, hampering practical application of NIPD in the clinic. Here, we have addressed this pitfall and devised a universal strategy for rapid NIPD of a prevalent mutation in the Ashkenazi Jewish (AJ) population.
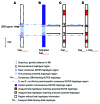
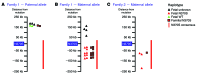
Methods: Pregnant AJ couples, carrying mutation(s) in GBA, which encodes acid β-glucosidase, were recruited at the SZMC Gaucher Clinic. Targeted next-generation sequencing of GBA-flanking SNPs was performed on peripheral blood samples from each couple, relevant mutation carrier family members, and unrelated individuals who are homozygotes for an AJ founder mutation. Allele-specific haplotypes were constructed based on linkage, and a consensus Gaucher disease-associated founder mutation-flanking haplotype was fine mapped. Together, these haplotypes were used for NIPD. All test results were validated by conventional prenatal or postnatal diagnostic methods.
Results: Ten parental alleles in eight unrelated fetuses were diagnosed successfully based on the noninvasive method developed in this study. The consensus mutation-flanking haplotype aided diagnosis for 6 of 9 founder mutation alleles.
Conclusions: The founder NIPD method developed and described here is rapid, economical, and readily adaptable for prenatal testing of prevalent autosomal recessive disease-causing mutations in an assortment of worldwide populations.
Funding: SZMC, Protalix Biotherapeutics Inc., and Centogene AG.
Figures



Similar articles
-
Reliable co-segregation analysis for prenatal diagnosis and heterozygote detection in Gaucher disease.Prenat Diagn. 1998 Mar;18(3):207-12. Prenat Diagn. 1998. PMID: 9556036
-
Gaucher disease: the origins of the Ashkenazi Jewish N370S and 84GG acid beta-glucosidase mutations.Am J Hum Genet. 2000 Jun;66(6):1821-32. doi: 10.1086/302946. Epub 2000 Apr 21. Am J Hum Genet. 2000. PMID: 10777718 Free PMC article.
-
Noninvasive prenatal diagnosis of β-thalassemia by relative haplotype dosage without analyzing proband.Mol Genet Genomic Med. 2019 Nov;7(11):e963. doi: 10.1002/mgg3.963. Epub 2019 Sep 30. Mol Genet Genomic Med. 2019. PMID: 31566929 Free PMC article.
-
Noninvasive approaches to prenatal diagnosis of hemoglobinopathies using fetal DNA in maternal plasma.Hematol Oncol Clin North Am. 2010 Dec;24(6):1179-86. doi: 10.1016/j.hoc.2010.08.007. Epub 2010 Sep 29. Hematol Oncol Clin North Am. 2010. PMID: 21075287 Review.
-
Non-invasive prenatal diagnosis by single molecule counting technologies.Trends Genet. 2009 Jul;25(7):324-31. doi: 10.1016/j.tig.2009.05.004. Epub 2009 Jun 18. Trends Genet. 2009. PMID: 19540612 Review.
Cited by
-
Noninvasive prenatal testing of α-thalassemia and β-thalassemia through population-based parental haplotyping.Genome Med. 2021 Feb 5;13(1):18. doi: 10.1186/s13073-021-00836-8. Genome Med. 2021. PMID: 33546747 Free PMC article.
-
Clinical evaluation of target capture sequencing technique for noninvasive prenatal diagnosis of β-thalassemia: A prospective case series.Medicine (Baltimore). 2025 Aug 22;104(34):e44014. doi: 10.1097/MD.0000000000044014. Medicine (Baltimore). 2025. PMID: 40859486 Free PMC article.
-
Non-Invasive Prenatal Diagnosis of Monogenic Disorders Through Bayesian- and Haplotype-Based Prediction of Fetal Genotype.Front Genet. 2022 Jul 1;13:911369. doi: 10.3389/fgene.2022.911369. eCollection 2022. Front Genet. 2022. PMID: 35846127 Free PMC article.
-
Fetal antisense oligonucleotide therapy for congenital deafness and vestibular dysfunction.Nucleic Acids Res. 2020 May 21;48(9):5065-5080. doi: 10.1093/nar/gkaa194. Nucleic Acids Res. 2020. PMID: 32249312 Free PMC article.
-
Noninvasive paternal exclusion testing for cystic fibrosis in the first five to eight weeks of gestation.Sci Rep. 2018 Oct 29;8(1):15941. doi: 10.1038/s41598-018-34396-6. Sci Rep. 2018. PMID: 30374031 Free PMC article.
References
-
- Lo YM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2(61): - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

