Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples
- PMID: 26426306
- PMCID: PMC4590409
- DOI: 10.3402/jom.v7.28934
Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples
Abstract
Background: Usefulness of next-generation sequencing (NGS) in assessing bacteria associated with oral squamous cell carcinoma (OSCC) has been undermined by inability to classify reads to the species level.
Objective: The purpose of this study was to develop a robust algorithm for species-level classification of NGS reads from oral samples and to pilot test it for profiling bacteria within OSCC tissues.
Methods: Bacterial 16S V1-V3 libraries were prepared from three OSCC DNA samples and sequenced using 454's FLX chemistry. High-quality, well-aligned, and non-chimeric reads ≥350 bp were classified using a novel, multi-stage algorithm that involves matching reads to reference sequences in revised versions of the Human Oral Microbiome Database (HOMD), HOMD extended (HOMDEXT), and Greengene Gold (GGG) at alignment coverage and percentage identity ≥98%, followed by assignment to species level based on top hit reference sequences. Priority was given to hits in HOMD, then HOMDEXT and finally GGG. Unmatched reads were subject to operational taxonomic unit analysis.
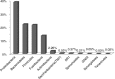
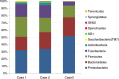
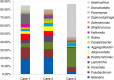
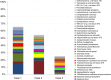
Results: Nearly, 92.8% of the reads were matched to updated-HOMD 13.2, 1.83% to trusted-HOMDEXT, and 1.36% to modified-GGG. Of all matched reads, 99.6% were classified to species level. A total of 228 species-level taxa were identified, representing 11 phyla; the most abundant were Proteobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Actinobacteria. Thirty-five species-level taxa were detected in all samples. On average, Prevotella oris, Neisseria flava, Neisseria flavescens/subflava, Fusobacterium nucleatum ss polymorphum, Aggregatibacter segnis, Streptococcus mitis, and Fusobacterium periodontium were the most abundant. Bacteroides fragilis, a species rarely isolated from the oral cavity, was detected in two samples.
Conclusion: This multi-stage algorithm maximizes the fraction of reads classified to the species level while ensuring reliable classification by giving priority to the human, oral reference set. Applying the algorithm to OSCC samples revealed high diversity. In addition to oral taxa, a number of human, non-oral taxa were also identified, some of which are rarely detected in the oral cavity.
Keywords: OSCC; bacteria; cancer; next-generation sequencing; pyrosequencing; taxonomy.
Figures


References
-
- Meurman JH. Oral microbiota and cancer. J Oral Microbiol. 2010;2 5195, doi: http://dx.doi.org/10.3402/jom.v2i0.5195. - DOI - PMC - PubMed
-
- Peter S, Beglinger C. Helicobacter pylori and gastric cancer: the causal relationship. Digestion. 2007;75:25–35. - PubMed
-
- Markowska J, Fischer N, Markowski M, Nalewaj J. The role of Chlamydia trachomatis infection in the development of cervical neoplasia and carcinoma. Med Wieku Rozwoj. 2005;9:83–6. - PubMed
-
- Nagaraja V, Eslick GD. Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment Pharmacol Ther. 2014;39:745–50. - PubMed
-
- Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–6. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
