A common promoter hypomethylation signature in invasive breast, liver and prostate cancer cell lines reveals novel targets involved in cancer invasiveness
- PMID: 26427334
- PMCID: PMC4741763
- DOI: 10.18632/oncotarget.5291
A common promoter hypomethylation signature in invasive breast, liver and prostate cancer cell lines reveals novel targets involved in cancer invasiveness
Abstract
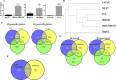
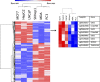
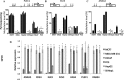
Cancer invasion and metastasis is the most morbid aspect of cancer and is governed by different cellular mechanisms than those driving the deregulated growth of tumors. We addressed here the question of whether a common DNA methylation signature of invasion exists in cancer cells from different origins that differentiates invasive from non-invasive cells. We identified a common DNA methylation signature consisting of hyper- and hypomethylation and determined the overlap of differences in DNA methylation with differences in mRNA expression using expression array analyses. A pathway analysis reveals that the hypomethylation signature includes some of the major pathways that were previously implicated in cancer migration and invasion such as TGF beta and ERBB2 triggered pathways. The relevance of these hypomethylation events in human tumors was validated by identification of the signature in several publicly available databases of human tumor transcriptomes. We shortlisted novel invasion promoting candidates and tested the role of four genes in cellular invasiveness from the list C11orf68, G0S2, SHISA2 and TMEM156 in invasiveness using siRNA depletion. Importantly these genes are upregulated in human cancer specimens as determined by immunostaining of human normal and cancer breast, liver and prostate tissue arrays. Since these genes are activated in cancer they constitute a group of targets for specific pharmacological inhibitors of cancer invasiveness.
Summary: Our study provides evidence that common DNA hypomethylation signature exists between cancer cells derived from different tissues, pointing to a common mechanism of cancer invasiveness in cancer cells from different origins that could serve as drug targets.
Keywords: DNA methylation; drug targets; epigenetics; hypomethylation; invasiveness.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- Graff J.R, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–9. - PubMed
-
- Herman J.G, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–30. - PubMed
-
- Pakneshan P, et al. Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J Biol Chem. 2004;279:31735–44. - PubMed
-
- Stefanska B, et al. Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res. 2011;71:5891–903. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

