Transformation of enriched mammary cell populations with polyomavirus middle T antigen influences tumor subtype and metastatic potential
- PMID: 26429062
- PMCID: PMC4589945
- DOI: 10.1186/s13058-015-0641-9
Transformation of enriched mammary cell populations with polyomavirus middle T antigen influences tumor subtype and metastatic potential
Abstract
Introduction: Breast cancer exhibits significant molecular, histological, and pathological diversity. Factors that impact this heterogeneity are poorly understood; however, transformation of distinct normal cell populations of the breast may generate different tumor phenotypes. Our previous study demonstrated that the polyomavirus middle T antigen (PyMT) oncogene can establish diverse tumor subtypes when broadly expressed within mouse mammary epithelial cells. In the present study, we assessed the molecular, histological, and metastatic outcomes in distinct mammary cell populations transformed with the PyMT gene.
Methods: Isolated mouse mammary epithelial cells were transduced with a lentivirus encoding PyMT during an overnight infection and then sorted into hormone receptor-positive luminal (CD133+), hormone receptor-negative luminal (CD133-), basal, and stem cell populations using the cell surface markers CD24, CD49f, and CD133. Each population was subsequently transplanted into syngeneic cleared mouse mammary fat pads to generate tumors. Tumors were classified by histology, estrogen receptor status, molecular subtype, and metastatic potential to investigate whether transformation of different enriched populations affects tumor phenotype.
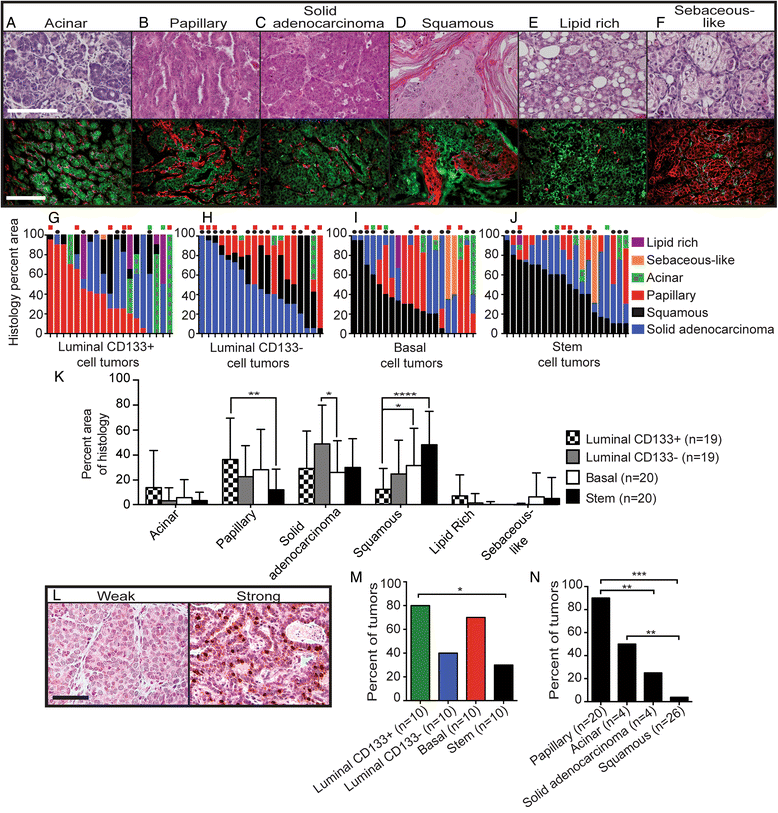
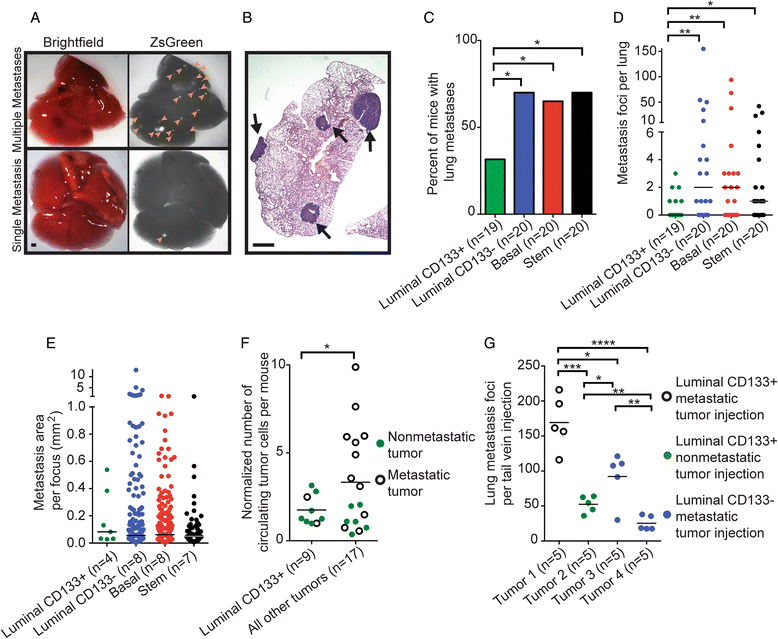
Results: Although enriched mammary epithelial cell populations showed no difference in either the ability to form tumors or tumor latency, differences in prevalence of solid adenocarcinomas and squamous, papillary, and sebaceous-like tumors were observed. In particular, squamous metaplasia was observed more frequently in tumors derived from basal and stem cells than in luminal cells. Interestingly, both molecularly basal and luminal tumors developed from luminal CD133+, basal, and stem cell populations; however, luminal CD133- cells gave rise exclusively to molecularly basal tumors. Tumors arising from the luminal CD133-, basal, and stem cell populations were highly metastatic; however, luminal CD133+ cells generated tumors that were significantly less metastatic, possibly due to an inability of these tumor cells to escape the primary tumor site.
Conclusions: Expression of PyMT within different mammary cell populations influences tumor histology, molecular subtype, and metastatic potential. The data demonstrate that luminal CD133+ cells give rise to less metastatic tumors, luminal CD133- cells preferentially establish basal tumors, and the cell of origin for squamous metaplasia likely resides in the basal and stem cell populations.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

