TumorTracer: a method to identify the tissue of origin from the somatic mutations of a tumor specimen
- PMID: 26429708
- PMCID: PMC4590711
- DOI: 10.1186/s12920-015-0130-0
TumorTracer: a method to identify the tissue of origin from the somatic mutations of a tumor specimen
Abstract
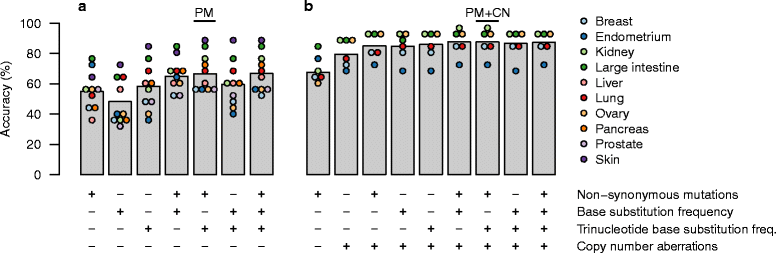
Background: A substantial proportion of cancer cases present with a metastatic tumor and require further testing to determine the primary site; many of these are never fully diagnosed and remain cancer of unknown primary origin (CUP). It has been previously demonstrated that the somatic point mutations detected in a tumor can be used to identify its site of origin with limited accuracy. We hypothesized that higher accuracy could be achieved by a classification algorithm based on the following feature sets: 1) the number of nonsynonymous point mutations in a set of 232 specific cancer-associated genes, 2) frequencies of the 96 classes of single-nucleotide substitution determined by the flanking bases, and 3) copy number profiles, if available.
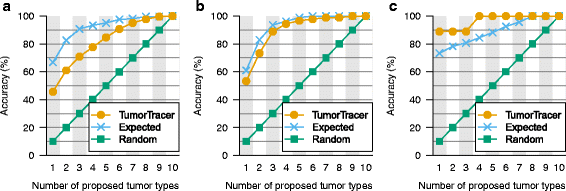
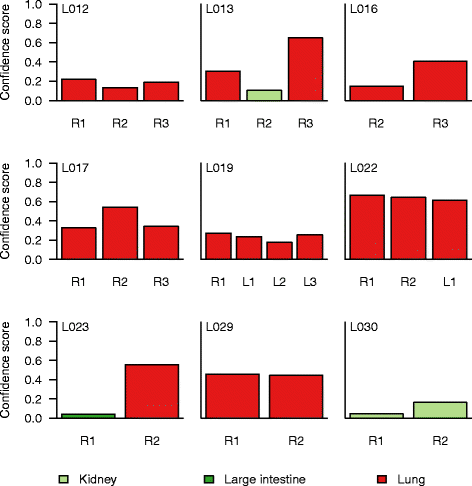
Methods: We used publicly available somatic mutation data from the COSMIC database to train random forest classifiers to distinguish among those tissues of origin for which sufficient data was available. We selected feature sets using cross-validation and then derived two final classifiers (with or without copy number profiles) using 80 % of the available tumors. We evaluated the accuracy using the remaining 20 %. For further validation, we assessed accuracy of the without-copy-number classifier on three independent data sets: 1669 newly available public tumors of various types, a cohort of 91 breast metastases, and a set of 24 specimens from 9 lung cancer patients subjected to multiregion sequencing.
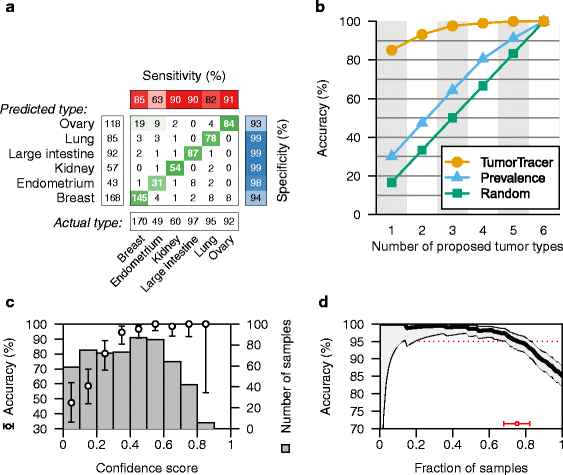
Results: The cross-validation accuracy was highest when all three types of information were used. On the left-out COSMIC data not used for training, we achieved a classification accuracy of 85 % across 6 primary sites (with copy numbers), and 69 % across 10 primary sites (without copy numbers). Importantly, a derived confidence score could distinguish tumors that could be identified with 95 % accuracy (32 %/75 % of tumors with/without copy numbers) from those that were less certain. Accuracy in the independent data sets was 46 %, 53 % and 89 % respectively, similar to the accuracy expected from the training data.
Conclusions: Identification of primary site from point mutation and/or copy number data may be accurate enough to aid clinical diagnosis of cancers of unknown primary origin.
Figures


References
-
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

