Expression Divergence of Chemosensory Genes between Drosophila sechellia and Its Sibling Species and Its Implications for Host Shift
- PMID: 26430061
- PMCID: PMC4684695
- DOI: 10.1093/gbe/evv183
Expression Divergence of Chemosensory Genes between Drosophila sechellia and Its Sibling Species and Its Implications for Host Shift
Abstract
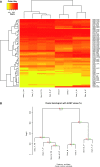
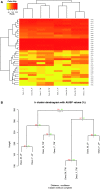
Drosophila sechellia relies exclusively on the fruits of Morinda citrifolia, which are toxic to most insects, including its sibling species Drosophila melanogaster and Drosophila simulans. Although several odorant binding protein (Obp) genes and olfactory receptor (Or) genes have been suggested to be associated with the D. sechellia host shift, a broad view of how chemosensory genes have contributed to this shift is still lacking. We therefore studied the transcriptomes of antennae, the main organ responsible for detecting food resource and oviposition, of D. sechellia and its two sibling species. We wanted to know whether gene expression, particularly chemosensory genes, has diverged between D. sechellia and its two sibling species. Using a very stringent definition of differential gene expression, we found a higher percentage of chemosensory genes differentially expressed in the D. sechellia lineage (7.8%) than in the D. simulans lineage (5.4%); for upregulated chemosensory genes, the percentages were 8.8% in D. sechellia and 5.2% in D. simulans. Interestingly, Obp50a exhibited the highest upregulation, an approximately 100-fold increase, and Or85c--previously reported to be a larva-specific gene--showed approximately 20-fold upregulation in D. sechellia. Furthermore, Ir84a (ionotropic receptor 84a), which has been proposed to be associated with male courtship behavior, was significantly upregulated in D. sechellia. We also found expression divergence in most of the chemosensory gene families between D. sechellia and the two sibling species. Our observations suggest that the host shift of D. sechellia was associated with the enrichment of differentially expressed, particularly upregulated, chemosensory genes.
Keywords: Drosophila sechellia; RNA-seq; antennal transcriptome; chemosensory genes; host shift.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Bassett AR, Liu J-LL. 2014. CRISPR/Cas9 and genome editing in Drosophila. J Genet Genomics. 41:7–19. - PubMed
-
- Clyne PJ, et al. 1999. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22:327–338. - PubMed
-
- Clyne PJ, Warr CG, Carlson JR. 2000. Candidate taste receptors in Drosophila. Science 287:1830–1834. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

