SRSF1-Regulated Alternative Splicing in Breast Cancer
- PMID: 26431027
- PMCID: PMC4597910
- DOI: 10.1016/j.molcel.2015.09.005
SRSF1-Regulated Alternative Splicing in Breast Cancer
Abstract
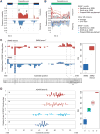
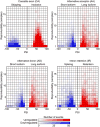
Splicing factor SRSF1 is upregulated in human breast tumors, and its overexpression promotes transformation of mammary cells. Using RNA-seq, we identified SRSF1-regulated alternative splicing (AS) targets in organotypic three-dimensional MCF-10A cell cultures that mimic a context relevant to breast cancer. We identified and validated hundreds of endogenous SRSF1-regulated AS events. De novo discovery of the SRSF1 binding motif reconciled discrepancies in previous motif analyses. Using a Bayesian model, we determined positional effects of SRSF1 binding on cassette exons: binding close to the 5' splice site generally promoted exon inclusion, whereas binding near the 3' splice site promoted either exon skipping or inclusion. Finally, we identified SRSF1-regulated AS events deregulated in human tumors; overexpressing one such isoform, exon-9-included CASC4, increased acinar size and proliferation, and decreased apoptosis, partially recapitulating SRSF1's oncogenic effects. Thus, we uncovered SRSF1 positive and negative regulatory mechanisms, and oncogenic AS events that represent potential targets for therapeutics development.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




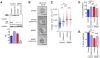
References
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

