Dynamics and Cell-Type Specificity of the DNA Double-Strand Break Repair Protein RecN in the Developmental Cyanobacterium Anabaena sp. Strain PCC 7120
- PMID: 26431054
- PMCID: PMC4592062
- DOI: 10.1371/journal.pone.0139362
Dynamics and Cell-Type Specificity of the DNA Double-Strand Break Repair Protein RecN in the Developmental Cyanobacterium Anabaena sp. Strain PCC 7120
Abstract
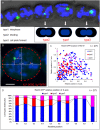
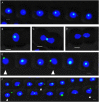
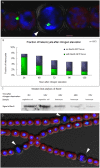
DNA replication and repair are two fundamental processes required in life proliferation and cellular defense and some common proteins are involved in both processes. The filamentous cyanobacterium Anabaena sp. strain PCC 7120 is capable of forming heterocysts for N2 fixation in the absence of a combined-nitrogen source. This developmental process is intimately linked to cell cycle control. In this study, we investigated the localization of the DNA double-strand break repair protein RecN during key cellular events, such as chromosome damaging, cell division, and heterocyst differentiation. Treatment by a drug causing DNA double-strand breaks (DSBs) induced reorganization of the RecN focus preferentially towards the mid-cell position. RecN-GFP was absent in most mature heterocysts. Furthermore, our results showed that HetR, a central player in heterocyst development, was involved in the proper positioning and distribution of RecN-GFP. These results showed the dynamics of RecN in DSB repair and suggested a differential regulation of DNA DSB repair in vegetative cell and heterocysts. The absence of RecN in mature heterocysts is compatible with the terminal nature of these cells.
Conflict of interest statement
Figures



Similar articles
-
Heterocyst development and diazotrophic metabolism in terminal respiratory oxidase mutants of the cyanobacterium Anabaena sp. strain PCC 7120.J Bacteriol. 2007 Jun;189(12):4425-30. doi: 10.1128/JB.00220-07. Epub 2007 Apr 6. J Bacteriol. 2007. PMID: 17416650 Free PMC article.
-
Alr5068, a Low-Molecular-Weight protein tyrosine phosphatase, is involved in formation of the heterocysts polysaccharide layer in the cyanobacterium Anabaena sp. PCC 7120.Res Microbiol. 2013 Oct;164(8):875-85. doi: 10.1016/j.resmic.2013.05.003. Epub 2013 Jul 1. Res Microbiol. 2013. PMID: 23827083
-
Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids.Mol Microbiol. 2004 Jun;52(6):1627-39. doi: 10.1111/j.1365-2958.2004.04102.x. Mol Microbiol. 2004. PMID: 15186413
-
Early steps of double-strand break repair in Bacillus subtilis.DNA Repair (Amst). 2013 Mar 1;12(3):162-76. doi: 10.1016/j.dnarep.2012.12.005. Epub 2013 Feb 4. DNA Repair (Amst). 2013. PMID: 23380520 Review.
-
Heterocyst differentiation and pattern formation in cyanobacteria: a chorus of signals.Mol Microbiol. 2006 Jan;59(2):367-75. doi: 10.1111/j.1365-2958.2005.04979.x. Mol Microbiol. 2006. PMID: 16390435 Review.
Cited by
-
Comparative Genomics of DNA Recombination and Repair in Cyanobacteria: Biotechnological Implications.Front Microbiol. 2016 Nov 9;7:1809. doi: 10.3389/fmicb.2016.01809. eCollection 2016. Front Microbiol. 2016. PMID: 27881980 Free PMC article. Review.
References
-
- Michel-Marks E, Courcelle CT, Korolev S, Courcelle J (2010) ATP binding, ATP hydrolysis, and protein dimerization are required for RecF to catalyze an early step in the processing and recovery of replication forks disrupted by DNA damage. J Mol Biol 401: 579–589. 10.1016/j.jmb.2010.06.013 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

