Host-Associated Metagenomics: A Guide to Generating Infectious RNA Viromes
- PMID: 26431175
- PMCID: PMC4592258
- DOI: 10.1371/journal.pone.0139810
Host-Associated Metagenomics: A Guide to Generating Infectious RNA Viromes
Expression of concern in
-
Expression of Concern: Host-Associated Metagenomics: A Guide to Generating Infectious RNA Viromes.PLoS One. 2022 Dec 13;17(12):e0279054. doi: 10.1371/journal.pone.0279054. eCollection 2022. PLoS One. 2022. PMID: 36512579 Free PMC article. No abstract available.
Abstract
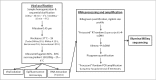
Background: Metagenomic analyses have been widely used in the last decade to describe viral communities in various environments or to identify the etiology of human, animal, and plant pathologies. Here, we present a simple and standardized protocol that allows for the purification and sequencing of RNA viromes from complex biological samples with an important reduction of host DNA and RNA contaminants, while preserving the infectivity of viral particles.
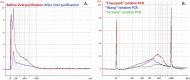
Principal findings: We evaluated different viral purification steps, random reverse transcriptions and sequence-independent amplifications of a pool of representative RNA viruses. Viruses remained infectious after the purification process. We then validated the protocol by sequencing the RNA virome of human body lice engorged in vitro with artificially contaminated human blood. The full genomes of the most abundant viruses absorbed by the lice during the blood meal were successfully sequenced. Interestingly, random amplifications differed in the genome coverage of segmented RNA viruses. Moreover, the majority of reads were taxonomically identified, and only 7-15% of all reads were classified as "unknown", depending on the random amplification method.
Conclusion: The protocol reported here could easily be applied to generate RNA viral metagenomes from complex biological samples of different origins. Our protocol allows further virological characterizations of the described viral communities because it preserves the infectivity of viral particles and allows for the isolation of viruses.
Conflict of interest statement
Figures

References
-
- Suttle CA. Viruses in the sea (2005) Nature 437: 356–361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

