Biofilm Formation Mechanisms of Pseudomonas aeruginosa Predicted via Genome-Scale Kinetic Models of Bacterial Metabolism
- PMID: 26431398
- PMCID: PMC4592021
- DOI: 10.1371/journal.pcbi.1004452
Biofilm Formation Mechanisms of Pseudomonas aeruginosa Predicted via Genome-Scale Kinetic Models of Bacterial Metabolism
Abstract
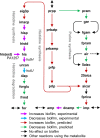
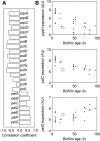
A hallmark of Pseudomonas aeruginosa is its ability to establish biofilm-based infections that are difficult to eradicate. Biofilms are less susceptible to host inflammatory and immune responses and have higher antibiotic tolerance than free-living planktonic cells. Developing treatments against biofilms requires an understanding of bacterial biofilm-specific physiological traits. Research efforts have started to elucidate the intricate mechanisms underlying biofilm development. However, many aspects of these mechanisms are still poorly understood. Here, we addressed questions regarding biofilm metabolism using a genome-scale kinetic model of the P. aeruginosa metabolic network and gene expression profiles. Specifically, we computed metabolite concentration differences between known mutants with altered biofilm formation and the wild-type strain to predict drug targets against P. aeruginosa biofilms. We also simulated the altered metabolism driven by gene expression changes between biofilm and stationary growth-phase planktonic cultures. Our analysis suggests that the synthesis of important biofilm-related molecules, such as the quorum-sensing molecule Pseudomonas quinolone signal and the exopolysaccharide Psl, is regulated not only through the expression of genes in their own synthesis pathway, but also through the biofilm-specific expression of genes in pathways competing for precursors to these molecules. Finally, we investigated why mutants defective in anthranilate degradation have an impaired ability to form biofilms. Alternative to a previous hypothesis that this biofilm reduction is caused by a decrease in energy production, we proposed that the dysregulation of the synthesis of secondary metabolites derived from anthranilate and chorismate is what impaired the biofilms of these mutants. Notably, these insights generated through our kinetic model-based approach are not accessible from previous constraint-based model analyses of P. aeruginosa biofilm metabolism. Our simulation results showed that plausible, non-intuitive explanations of difficult-to-interpret experimental observations could be generated by integrating genome-scale kinetic models with gene expression profiles.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Pseudomonas aeruginosa Biofilm Antibiotic Resistance Gene ndvB Expression Requires the RpoS Stationary-Phase Sigma Factor.Appl Environ Microbiol. 2018 Mar 19;84(7):e02762-17. doi: 10.1128/AEM.02762-17. Print 2018 Apr 1. Appl Environ Microbiol. 2018. PMID: 29352081 Free PMC article.
-
Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation.J Bacteriol. 2001 Mar;183(6):1990-6. doi: 10.1128/JB.183.6.1990-1996.2001. J Bacteriol. 2001. PMID: 11222597 Free PMC article.
-
PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes.Appl Microbiol Biotechnol. 2008 Feb;78(2):293-307. doi: 10.1007/s00253-007-1308-y. Epub 2007 Dec 22. Appl Microbiol Biotechnol. 2008. PMID: 18157527
-
Pseudomonas aeruginosa biofilm infections: from molecular biofilm biology to new treatment possibilities.APMIS Suppl. 2014 Dec;(138):1-51. doi: 10.1111/apm.12335. APMIS Suppl. 2014. PMID: 25399808 Review.
-
Biofilm dispersion in Pseudomonas aeruginosa.J Microbiol. 2016 Feb;54(2):71-85. doi: 10.1007/s12275-016-5528-7. Epub 2016 Feb 2. J Microbiol. 2016. PMID: 26832663 Review.
Cited by
-
Regulation of Pseudomonas aeruginosa-Mediated Neutrophil Extracellular Traps.Front Immunol. 2019 Jul 18;10:1670. doi: 10.3389/fimmu.2019.01670. eCollection 2019. Front Immunol. 2019. PMID: 31379861 Free PMC article.
-
Participation of Acyl-Coenzyme A Synthetase FadD4 of Pseudomonas aeruginosa PAO1 in Acyclic Terpene/Fatty Acid Assimilation and Virulence by Lipid A Modification.Front Microbiol. 2021 Nov 16;12:785112. doi: 10.3389/fmicb.2021.785112. eCollection 2021. Front Microbiol. 2021. PMID: 34867927 Free PMC article.
-
Quantitative analysis of Mycobacterium avium subsp. hominissuis proteome in response to antibiotics and during exposure to different environmental conditions.Clin Proteomics. 2019 Nov 13;16:39. doi: 10.1186/s12014-019-9260-2. eCollection 2019. Clin Proteomics. 2019. PMID: 31749666 Free PMC article.
-
Antibiotic resistance: Time of synthesis in a post-genomic age.Comput Struct Biotechnol J. 2021 May 21;19:3110-3124. doi: 10.1016/j.csbj.2021.05.034. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34141134 Free PMC article. Review.
-
Novel antimicrobial development using genome-scale metabolic model of Gram-negative pathogens: a review.J Antibiot (Tokyo). 2021 Feb;74(2):95-104. doi: 10.1038/s41429-020-00366-2. Epub 2020 Sep 8. J Antibiot (Tokyo). 2021. PMID: 32901119 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

