MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells
- PMID: 26431963
- PMCID: PMC4643329
- DOI: 10.1042/BSR20150194
MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells
Erratum in
-
Correction: MELK-T1, a small-molecule inhibitor of protein kinase MELK, decreases DNA-damage tolerance in proliferating cancer cells.Biosci Rep. 2018 Nov 30;38(6):BSR-20150194_COR. doi: 10.1042/BSR-20150194_COR. Print 2018 Dec 21. Biosci Rep. 2018. PMID: 30504271 Free PMC article. No abstract available.
Abstract
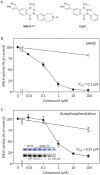
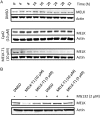
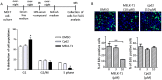
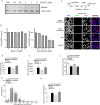
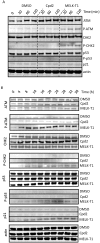
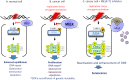
Maternal embryonic leucine zipper kinase (MELK), a serine/threonine protein kinase, has oncogenic properties and is overexpressed in many cancer cells. The oncogenic function of MELK is attributed to its capacity to disable critical cell-cycle checkpoints and reduce replication stress. Most functional studies have relied on the use of siRNA/shRNA-mediated gene silencing. In the present study, we have explored the biological function of MELK using MELK-T1, a novel and selective small-molecule inhibitor. Strikingly, MELK-T1 triggered a rapid and proteasome-dependent degradation of the MELK protein. Treatment of MCF-7 (Michigan Cancer Foundation-7) breast adenocarcinoma cells with MELK-T1 induced the accumulation of stalled replication forks and double-strand breaks that culminated in a replicative senescence phenotype. This phenotype correlated with a rapid and long-lasting ataxia telangiectasia-mutated (ATM) activation and phosphorylation of checkpoint kinase 2 (CHK2). Furthermore, MELK-T1 induced a strong phosphorylation of p53 (cellular tumour antigen p53), a prolonged up-regulation of p21 (cyclin-dependent kinase inhibitor 1) and a down-regulation of FOXM1 (Forkhead Box M1) target genes. Our data indicate that MELK is a key stimulator of proliferation by its ability to increase the threshold for DNA-damage tolerance (DDT). Thus, targeting MELK by the inhibition of both its catalytic activity and its protein stability might sensitize tumours to DNA-damaging agents or radiation therapy by lowering the DNA-damage threshold.
Keywords: chemical biology; deoxyribonucleic acid (DNA) damage response; maternal embryonic leucine zipper kinase (MELK) kinase; senescence; small molecule inhibitors.
© 2015 Authors.
Figures


References
-
- Lizcano J.M., Göransson O., Toth R., Deak M., Morrice N.A., Boudeau J., Hawley S.A., Udd L., Mäkelä T.P., Hardie D.G., Alessi D.R. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

