Subcellular Distribution of NAD+ between Cytosol and Mitochondria Determines the Metabolic Profile of Human Cells
- PMID: 26432643
- PMCID: PMC4646015
- DOI: 10.1074/jbc.M115.654129
Subcellular Distribution of NAD+ between Cytosol and Mitochondria Determines the Metabolic Profile of Human Cells
Abstract
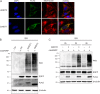
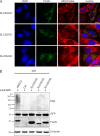
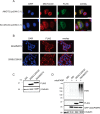
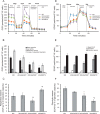
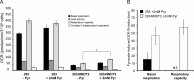
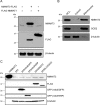
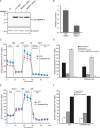
The mitochondrial NAD pool is particularly important for the maintenance of vital cellular functions. Although at least in some fungi and plants, mitochondrial NAD is imported from the cytosol by carrier proteins, in mammals, the mechanism of how this organellar pool is generated has remained obscure. A transporter mediating NAD import into mammalian mitochondria has not been identified. In contrast, human recombinant NMNAT3 localizes to the mitochondrial matrix and is able to catalyze NAD(+) biosynthesis in vitro. However, whether the endogenous NMNAT3 protein is functionally effective at generating NAD(+) in mitochondria of intact human cells still remains to be demonstrated. To modulate mitochondrial NAD(+) content, we have expressed plant and yeast mitochondrial NAD(+) carriers in human cells and observed a profound increase in mitochondrial NAD(+). None of the closest human homologs of these carriers had any detectable effect on mitochondrial NAD(+) content. Surprisingly, constitutive redistribution of NAD(+) from the cytosol to the mitochondria by stable expression of the Arabidopsis thaliana mitochondrial NAD(+) transporter NDT2 in HEK293 cells resulted in dramatic growth retardation and a metabolic shift from oxidative phosphorylation to glycolysis, despite the elevated mitochondrial NAD(+) levels. These results suggest that a mitochondrial NAD(+) transporter, similar to the known one from A. thaliana, is likely absent and could even be harmful in human cells. We provide further support for the alternative possibility, namely intramitochondrial NAD(+) synthesis, by demonstrating the presence of endogenous NMNAT3 in the mitochondria of human cells.
Keywords: NAD transport; NMNAT3; cell compartmentalization; glycolysis; metabolism; mitochondria; respiration.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Koch-Nolte F., Haag F., Guse A. H., Lund F., and Ziegler M. (2009) Emerging roles of NAD+ and its metabolites in cell signaling. Sci. Signal. 2, mr1. - PubMed
-
- Szendroedi J., Phielix E., and Roden M. (2012) The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 8, 92–103 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

