A Phylogenetically Conserved Group of Nuclear Factor-Y Transcription Factors Interact to Control Nodulation in Legumes
- PMID: 26432878
- PMCID: PMC4677902
- DOI: 10.1104/pp.15.01144
A Phylogenetically Conserved Group of Nuclear Factor-Y Transcription Factors Interact to Control Nodulation in Legumes
Erratum in
-
CORRECTION: Vol. 169: 2761-2773, 2015.Plant Physiol. 2017 Apr;173(4):2413. doi: 10.1104/pp.17.00334. Plant Physiol. 2017. PMID: 28356545 Free PMC article. No abstract available.
Abstract
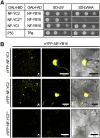
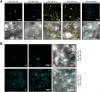
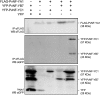
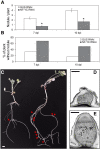
The endosymbiotic association between legumes and soil bacteria called rhizobia leads to the formation of a new root-derived organ called the nodule in which differentiated bacteria convert atmospheric nitrogen into a form that can be assimilated by the host plant. Successful root infection by rhizobia and nodule organogenesis require the activation of symbiotic genes that are controlled by a set of transcription factors (TFs). We recently identified Medicago truncatula nuclear factor-YA1 (MtNF-YA1) and MtNF-YA2 as two M. truncatula TFs playing a central role during key steps of the Sinorhizobium meliloti-M. truncatula symbiotic interaction. NF-YA TFs interact with NF-YB and NF-YC subunits to regulate target genes containing the CCAAT box consensus sequence. In this study, using a yeast two-hybrid screen approach, we identified the NF-YB and NF-YC subunits able to interact with MtNF-YA1 and MtNF-YA2. In yeast (Saccharomyces cerevisiae) and in planta, we further demonstrated by both coimmunoprecipitation and bimolecular fluorescence complementation that these NF-YA, -B, and -C subunits interact and form a stable NF-Y heterotrimeric complex. Reverse genetic and chromatin immunoprecipitation-PCR approaches revealed the importance of these newly identified NF-YB and NF-YC subunits for rhizobial symbiosis and binding to the promoter of MtERN1 (for Ethylene Responsive factor required for Nodulation), a direct target gene of MtNF-YA1 and MtNF-YA2. Finally, we verified that a similar trimer is formed in planta by the common bean (Phaseolus vulgaris) NF-Y subunits, revealing the existence of evolutionary conserved NF-Y protein complexes to control nodulation in leguminous plants. This sheds light on the process whereby an ancient heterotrimeric TF mainly controlling cell division in animals has acquired specialized functions in plants.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures



References
-
- Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN (2003) The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res 63: 8167–8172 - PubMed
-
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GED, et al. (2014) The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

