Physiological Functions of the COPI Complex in Higher Plants
- PMID: 26434491
- PMCID: PMC4625068
- DOI: 10.14348/molcells.2015.0115
Physiological Functions of the COPI Complex in Higher Plants
Abstract
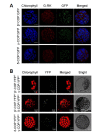
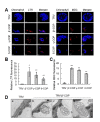
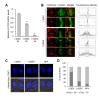
COPI vesicles are essential to the retrograde transport of proteins in the early secretory pathway. The COPI coatomer complex consists of seven subunits, termed α-, β-, β'-, γ-, δ-, ε-, and ζ-COP, in yeast and mammals. Plant genomes have homologs of these subunits, but the essentiality of their cellular functions has hampered the functional characterization of the subunit genes in plants. Here we have employed virus-induced gene silencing (VIGS) and dexamethasone (DEX)-inducible RNAi of the COPI subunit genes to study the in vivo functions of the COPI coatomer complex in plants. The β'-, γ-, and δ-COP subunits localized to the Golgi as GFP-fusion proteins and interacted with each other in the Golgi. Silencing of β'-, γ-, and δ-COP by VIGS resulted in growth arrest and acute plant death in Nicotiana benthamiana, with the affected leaf cells exhibiting morphological markers of programmed cell death. Depletion of the COPI subunits resulted in disruption of the Golgi structure and accumulation of autolysosome-like structures in earlier stages of gene silencing. In tobacco BY-2 cells, DEX-inducible RNAi of β'-COP caused aberrant cell plate formation during cytokinesis. Collectively, these results suggest that COPI vesicles are essential to plant growth and survival by maintaining the Golgi apparatus and modulating cell plate formation.
Keywords: COPI vesicle; Golgi localization; autophagy; cell death; cell plate formation; virus-induced gene silencing.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

