Bacterial danger sensing
- PMID: 26434507
- PMCID: PMC4658316
- DOI: 10.1016/j.jmb.2015.09.018
Bacterial danger sensing
Abstract
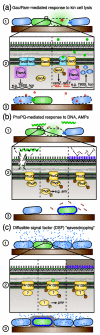
Here we propose that bacteria detect and respond to threats posed by other bacteria via an innate immune-like process that we term danger sensing. We find support for this contention by reexamining existing literature from the perspective that intermicrobial antagonism, not opportunistic pathogenesis, is the major evolutionary force shaping the defensive behaviors of most bacteria. We conclude that many bacteria possess danger sensing pathways composed of a danger signal receptor and corresponding signal transduction mechanism that regulate pathways important for survival in the presence of the perceived competitor.
Keywords: Gac/Rsm; competence; interbacterial; subinhibitory antibiotics; type VI secretion.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
References
-
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. - PubMed
-
- Shi Y, Evans JE, Rock KL. Molecular identification of a danger signal that alerts the immune system to dying cells. Nature. 2003;425:516–21. - PubMed
-
- Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

